INFERNO: inferring the molecular mechanisms of noncoding genetic variants
- PMID: 30113658
- PMCID: PMC6158604
- DOI: 10.1093/nar/gky686
INFERNO: inferring the molecular mechanisms of noncoding genetic variants
Abstract
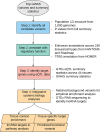
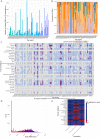
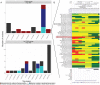
The majority of variants identified by genome-wide association studies (GWAS) reside in the noncoding genome, affecting regulatory elements including transcriptional enhancers. However, characterizing their effects requires the integration of GWAS results with context-specific regulatory activity and linkage disequilibrium annotations to identify causal variants underlying noncoding association signals and the regulatory elements, tissue contexts, and target genes they affect. We propose INFERNO, a novel method which integrates hundreds of functional genomics datasets spanning enhancer activity, transcription factor binding sites, and expression quantitative trait loci with GWAS summary statistics. INFERNO includes novel statistical methods to quantify empirical enrichments of tissue-specific enhancer overlap and to identify co-regulatory networks of dysregulated long noncoding RNAs (lncRNAs). We applied INFERNO to two large GWAS studies. For schizophrenia (36,989 cases, 113,075 controls), INFERNO identified putatively causal variants affecting brain enhancers for known schizophrenia-related genes. For inflammatory bowel disease (IBD) (12,882 cases, 21,770 controls), INFERNO found enrichments of immune and digestive enhancers and lncRNAs involved in regulation of the adaptive immune response. In summary, INFERNO comprehensively infers the molecular mechanisms of causal noncoding variants, providing a sensitive hypothesis generation method for post-GWAS analysis. The software is available as an open source pipeline and a web server.
Figures

Similar articles
-
Inferring the Molecular Mechanisms of Noncoding Alzheimer's Disease-Associated Genetic Variants.J Alzheimers Dis. 2019;72(1):301-318. doi: 10.3233/JAD-190568. J Alzheimers Dis. 2019. PMID: 31561366 Free PMC article.
-
Using INFERNO to Infer the Molecular Mechanisms Underlying Noncoding Genetic Associations.Methods Mol Biol. 2021;2254:73-91. doi: 10.1007/978-1-0716-1158-6_6. Methods Mol Biol. 2021. PMID: 33326071
-
Genome-wide enhancer maps link risk variants to disease genes.Nature. 2021 May;593(7858):238-243. doi: 10.1038/s41586-021-03446-x. Epub 2021 Apr 7. Nature. 2021. PMID: 33828297 Free PMC article.
-
Functional Implications of Intergenic GWAS SNPs in Immune-Related LncRNAs.Adv Exp Med Biol. 2022;1363:147-160. doi: 10.1007/978-3-030-92034-0_8. Adv Exp Med Biol. 2022. PMID: 35220569 Review.
-
Identifying causal variants and genes using functional genomics in specialized cell types and contexts.Hum Genet. 2020 Jan;139(1):95-102. doi: 10.1007/s00439-019-02044-2. Epub 2019 Jul 17. Hum Genet. 2020. PMID: 31317254 Free PMC article. Review.
Cited by
-
Unfolding the genotype-to-phenotype black box of cardiovascular diseases through cross-scale modeling.iScience. 2022 Jul 20;25(8):104790. doi: 10.1016/j.isci.2022.104790. eCollection 2022 Aug 19. iScience. 2022. PMID: 35992073 Free PMC article.
-
QBiC-Pred: quantitative predictions of transcription factor binding changes due to sequence variants.Nucleic Acids Res. 2019 Jul 2;47(W1):W127-W135. doi: 10.1093/nar/gkz363. Nucleic Acids Res. 2019. PMID: 31114870 Free PMC article.
-
Genetic meta-analysis of diagnosed Alzheimer's disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing.Nat Genet. 2019 Mar;51(3):414-430. doi: 10.1038/s41588-019-0358-2. Epub 2019 Feb 28. Nat Genet. 2019. PMID: 30820047 Free PMC article.
-
The goldmine of GWAS summary statistics: a systematic review of methods and tools.BioData Min. 2024 Sep 5;17(1):31. doi: 10.1186/s13040-024-00385-x. BioData Min. 2024. PMID: 39238044 Free PMC article.
-
Scalable approaches for functional analyses of whole-genome sequencing non-coding variants.Hum Mol Genet. 2022 Oct 20;31(R1):R62-R72. doi: 10.1093/hmg/ddac191. Hum Mol Genet. 2022. PMID: 35943817 Free PMC article. Review.
References
-
- Evangelou E., Ioannidis J.P.A.. Meta-analysis methods for genome-wide association studies and beyond. Nat. Rev. Genet. 2013; 14:379–389. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

