Gene essentiality landscape and druggable oncogenic dependencies in herpesviral primary effusion lymphoma
- PMID: 30111820
- PMCID: PMC6093911
- DOI: 10.1038/s41467-018-05506-9
Gene essentiality landscape and druggable oncogenic dependencies in herpesviral primary effusion lymphoma
Abstract
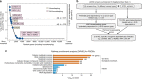
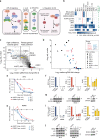
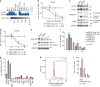
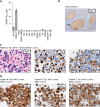
Primary effusion lymphoma (PEL) is caused by Kaposi's sarcoma-associated herpesvirus. Our understanding of PEL is poor and therefore treatment strategies are lacking. To address this need, we conducted genome-wide CRISPR/Cas9 knockout screens in eight PEL cell lines. Integration with data from unrelated cancers identifies 210 genes as PEL-specific oncogenic dependencies. Genetic requirements of PEL cell lines are largely independent of Epstein-Barr virus co-infection. Genes of the NF-κB pathway are individually non-essential. Instead, we demonstrate requirements for IRF4 and MDM2. PEL cell lines depend on cellular cyclin D2 and c-FLIP despite expression of viral homologs. Moreover, PEL cell lines are addicted to high levels of MCL1 expression, which are also evident in PEL tumors. Strong dependencies on cyclin D2 and MCL1 render PEL cell lines highly sensitive to palbociclib and S63845. In summary, this work comprehensively identifies genetic dependencies in PEL cell lines and identifies novel strategies for therapeutic intervention.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Expression Ratios of the Antiapoptotic BCL2 Family Members Dictate the Selective Addiction of Kaposi's Sarcoma-Associated Herpesvirus-Transformed Primary Effusion Lymphoma Cell Lines to MCL1.J Virol. 2022 Dec 14;96(23):e0136022. doi: 10.1128/jvi.01360-22. Epub 2022 Nov 23. J Virol. 2022. PMID: 36416587 Free PMC article.
-
Druggable host gene dependencies in primary effusion lymphoma.Curr Opin Virol. 2022 Oct;56:101270. doi: 10.1016/j.coviro.2022.101270. Epub 2022 Sep 28. Curr Opin Virol. 2022. PMID: 36182745 Free PMC article. Review.
-
Interleukin-1 Receptor-Associated Kinase (IRAK) Signaling in Kaposi Sarcoma-Associated Herpesvirus-Induced Primary Effusion Lymphoma.J Virol. 2020 May 4;94(10):e02123-19. doi: 10.1128/JVI.02123-19. Print 2020 May 4. J Virol. 2020. PMID: 32161170 Free PMC article.
-
Altering the Anti-inflammatory Lipoxin Microenvironment: a New Insight into Kaposi's Sarcoma-Associated Herpesvirus Pathogenesis.J Virol. 2016 Nov 28;90(24):11020-11031. doi: 10.1128/JVI.01491-16. Print 2016 Dec 15. J Virol. 2016. PMID: 27681120 Free PMC article.
-
[Replication Machinery of Kaposi's Sarcoma-associated Herpesvirus and Drug Discovery Research].Yakugaku Zasshi. 2019;139(1):69-73. doi: 10.1248/yakushi.18-00164-2. Yakugaku Zasshi. 2019. PMID: 30606932 Review. Japanese.
Cited by
-
The Mitochondrial Ubiquitin Ligase MARCHF5 Cooperates with MCL1 to Inhibit Apoptosis in KSHV-Transformed Primary Effusion Lymphoma Cell Lines.bioRxiv [Preprint]. 2024 Sep 24:2024.09.23.614413. doi: 10.1101/2024.09.23.614413. bioRxiv. 2024. PMID: 39386614 Free PMC article. Preprint.
-
CRISPR screens identify novel regulators of cFLIP dependency and ligand-independent, TRAIL-R1-mediated cell death.Cell Death Differ. 2023 May;30(5):1221-1234. doi: 10.1038/s41418-023-01133-0. Epub 2023 Feb 18. Cell Death Differ. 2023. PMID: 36801923 Free PMC article.
-
Transcription Factor IRF4 Dysfunction Affects the Immunosuppressive Function of Treg Cells in Patients with Primary Immune Thrombocytopenia.Biomed Res Int. 2019 Jul 10;2019:1050285. doi: 10.1155/2019/1050285. eCollection 2019. Biomed Res Int. 2019. PMID: 31380412 Free PMC article.
-
A virus-induced circular RNA maintains latent infection of Kaposi's sarcoma herpesvirus.Proc Natl Acad Sci U S A. 2023 Feb 7;120(6):e2212864120. doi: 10.1073/pnas.2212864120. Epub 2023 Feb 1. Proc Natl Acad Sci U S A. 2023. PMID: 36724259 Free PMC article.
-
CRISPR-Cas9 Screening of Kaposi's Sarcoma-Associated Herpesvirus-Transformed Cells Identifies XPO1 as a Vulnerable Target of Cancer Cells.mBio. 2019 May 14;10(3):e00866-19. doi: 10.1128/mBio.00866-19. mBio. 2019. PMID: 31088931 Free PMC article.
References
-
- Nador RG, et al. Primary effusion lymphoma: a distinct clinicopathologic entity associated with the Kaposi’s sarcoma-associated herpes virus. Blood. 1996;88:645–656. - PubMed
-
- Soulier J, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

