Notch Signaling Modulates Macrophage Polarization and Phagocytosis Through Direct Suppression of Signal Regulatory Protein α Expression
- PMID: 30105024
- PMCID: PMC6077186
- DOI: 10.3389/fimmu.2018.01744
Notch Signaling Modulates Macrophage Polarization and Phagocytosis Through Direct Suppression of Signal Regulatory Protein α Expression
Abstract
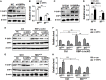
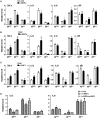
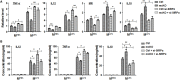
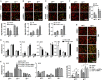
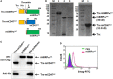
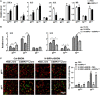
The Notch pathway plays critical roles in the development and functional modulation of myeloid cells. Previous studies have demonstrated that Notch activation promotes M1 polarization and phagocytosis of macrophages; however, the downstream molecular mechanisms mediating Notch signal remain elusive. In an attempt to identify Notch downstream targets in bone marrow-derived macrophages (BMDMs) using mass spectrometry, the signal regulatory protein α (SIRPα) appeared to respond to knockout of recombination signal-binding protein Jk (RBP-J), the critical transcription factor of Notch pathway, in macrophages. In this study, we validated that Notch activation could repress SIRPα expression likely via the Hes family co-repressors. SIRPα promoted macrophage M2 polarization, which was dependent on the interaction with CD47 and mediated by intracellular signaling through SHP-1. We provided evidence that Notch signal regulated macrophage polarization at least partially through SIRPα. Interestingly, Notch signal regulated macrophage phagocytosis of tumor cells through SIRPα but in a SHP-1-independent way. To access the translational value of our findings, we expressed the extracellular domains of the mouse SIRPα (mSIRPαext) to block the interaction between CD47 and SIRPα. We demonstrated that the soluble mSIRPαext polypeptides could promote M1 polarization and increase phagocytosis of tumor cells by macrophages. Taken together, our results provided new insights into the molecular mechanisms of notch-mediated macrophage polarization and further validated SIRPα as a target for tumor therapy through modulating macrophage polarization and phagocytosis.
Keywords: SHP-1; macrophages; notch; phagocytosis; polarization; signal regulatory protein α.
Figures

Similar articles
-
"Velcro" engineering of high affinity CD47 ectodomain as signal regulatory protein α (SIRPα) antagonists that enhance antibody-dependent cellular phagocytosis.J Biol Chem. 2015 May 15;290(20):12650-63. doi: 10.1074/jbc.M115.648220. Epub 2015 Apr 2. J Biol Chem. 2015. PMID: 25837251 Free PMC article.
-
[The extracellular segment of mouse soluble SIRPα enhances the phagocytosis of macrophages to L1210 leukemia cells].Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2018 Dec;34(12):1057-1062. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2018. PMID: 30626469 Chinese.
-
CD47-SIRPα Interactions Regulate Macrophage Uptake of Plasmodium falciparum-Infected Erythrocytes and Clearance of Malaria In Vivo.Infect Immun. 2016 Jun 23;84(7):2002-2011. doi: 10.1128/IAI.01426-15. Print 2016 Jul. Infect Immun. 2016. PMID: 27091932 Free PMC article.
-
The CD47-SIRPα signalling system: its physiological roles and therapeutic application.J Biochem. 2014 Jun;155(6):335-44. doi: 10.1093/jb/mvu017. Epub 2014 Mar 12. J Biochem. 2014. PMID: 24627525 Review.
-
Cancer immunotherapy targeting the CD47/SIRPα axis.Eur J Cancer. 2017 May;76:100-109. doi: 10.1016/j.ejca.2017.02.013. Epub 2017 Mar 10. Eur J Cancer. 2017. PMID: 28286286 Review.
Cited by
-
Linc00514 promotes breast cancer metastasis and M2 polarization of tumor-associated macrophages via Jagged1-mediated notch signaling pathway.J Exp Clin Cancer Res. 2020 Sep 17;39(1):191. doi: 10.1186/s13046-020-01676-x. J Exp Clin Cancer Res. 2020. PMID: 32943090 Free PMC article.
-
Vasoactive intestinal peptide blockade suppresses tumor growth by regulating macrophage polarization and function in CT26 tumor-bearing mice.Sci Rep. 2023 Jan 17;13(1):927. doi: 10.1038/s41598-023-28073-6. Sci Rep. 2023. PMID: 36650220 Free PMC article.
-
Effects of Metabolism on Macrophage Polarization Under Different Disease Backgrounds.Front Immunol. 2022 Jul 14;13:880286. doi: 10.3389/fimmu.2022.880286. eCollection 2022. Front Immunol. 2022. PMID: 35911719 Free PMC article. Review.
-
Macrophages in aseptic loosening: Characteristics, functions, and mechanisms.Front Immunol. 2023 Mar 8;14:1122057. doi: 10.3389/fimmu.2023.1122057. eCollection 2023. Front Immunol. 2023. PMID: 36969165 Free PMC article. Review.
-
Engineering Pancreatic Islets to Transiently Codisplay on Their Surface Thrombomodulin and CD47 Immunomodulatory Proteins as a Means of Mitigating Instant Blood-Mediated Inflammatory Reaction following Intraportal Transplantation.J Immunol. 2024 Jun 15;212(12):1971-1980. doi: 10.4049/jimmunol.2300743. J Immunol. 2024. PMID: 38709159
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

