Endogenous HIF2A reporter systems for high-throughput functional screening
- PMID: 30104738
- PMCID: PMC6089976
- DOI: 10.1038/s41598-018-30499-2
Endogenous HIF2A reporter systems for high-throughput functional screening
Abstract
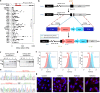
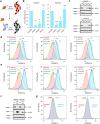
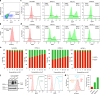
Tissue-specific transcriptional programs control most biological phenotypes, including disease states such as cancer. However, the molecular details underlying transcriptional specificity is largely unknown, hindering the development of therapeutic approaches. Here, we describe novel experimental reporter systems that allow interrogation of the endogenous expression of HIF2A, a critical driver of renal oncogenesis. Using a focused CRISPR-Cas9 library targeting chromatin regulators, we provide evidence that these reporter systems are compatible with high-throughput screening. Our data also suggests redundancy in the control of cancer type-specific transcriptional traits. Reporter systems such as those described here could facilitate large-scale mechanistic dissection of transcriptional programmes underlying cancer phenotypes, thus paving the way for novel therapeutic approaches.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Development of drug-inducible CRISPR-Cas9 systems for large-scale functional screening.BMC Genomics. 2019 Mar 19;20(1):225. doi: 10.1186/s12864-019-5601-9. BMC Genomics. 2019. PMID: 30890156 Free PMC article.
-
Establishment of a DGKθ Endogenous Promoter Luciferase Reporter HepG2 Cell Line for Studying the Transcriptional Regulation of DGKθ Gene.Appl Biochem Biotechnol. 2019 Apr;187(4):1344-1355. doi: 10.1007/s12010-018-2890-4. Epub 2018 Sep 19. Appl Biochem Biotechnol. 2019. PMID: 30229432
-
Generation of a novel HEK293 luciferase reporter cell line by CRISPR/Cas9-mediated site-specific integration in the genome to explore the transcriptional regulation of the PGRN gene.Bioengineered. 2019 Dec;10(1):98-107. doi: 10.1080/21655979.2019.1607126. Bioengineered. 2019. PMID: 31023186 Free PMC article.
-
High-throughput genetic screens using CRISPR-Cas9 system.Arch Pharm Res. 2018 Sep;41(9):875-884. doi: 10.1007/s12272-018-1029-z. Epub 2018 Apr 10. Arch Pharm Res. 2018. PMID: 29637495 Review.
-
Large scale control and programming of gene expression using CRISPR.Semin Cell Dev Biol. 2019 Dec;96:124-132. doi: 10.1016/j.semcdb.2019.05.013. Epub 2019 Jun 12. Semin Cell Dev Biol. 2019. PMID: 31181342 Review.
Cited by
-
The SWI/SNF complex member SMARCB1 supports lineage fidelity in kidney cancer.iScience. 2023 Jul 13;26(8):107360. doi: 10.1016/j.isci.2023.107360. eCollection 2023 Aug 18. iScience. 2023. PMID: 37554444 Free PMC article.
-
Targeting HIF-2α in the Tumor Microenvironment: Redefining the Role of HIF-2α for Solid Cancer Therapy.Cancers (Basel). 2022 Feb 28;14(5):1259. doi: 10.3390/cancers14051259. Cancers (Basel). 2022. PMID: 35267567 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

