Quantitative Determination of MAR Hydrolase Residue Specificity In Vitro by Tandem Mass Spectrometry
- PMID: 30097875
- PMCID: PMC6382470
- DOI: 10.1007/978-1-4939-8588-3_19
Quantitative Determination of MAR Hydrolase Residue Specificity In Vitro by Tandem Mass Spectrometry
Erratum in
-
Correction to: ADP-ribosylation and NAD+ Utilizing Enzymes.Methods Mol Biol. 2018;1813:C1. doi: 10.1007/978-1-4939-8588-3_27. Methods Mol Biol. 2018. PMID: 30488319 Free PMC article.
Abstract
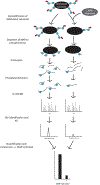
ADP-ribosylation is a posttranslational modification that involves the conjugation of monomers and polymers of the small molecule ADP-ribose onto amino acid side chains. A family of ADP-ribosyltransferases catalyzes the transfer of the ADP-ribose moiety of nicotinamide adenine dinucleotide (NAD+) onto a variety of amino acid side chains including aspartate, glutamate, lysine, arginine, cysteine, and serine. The monomeric form of the modification mono(ADP-ribosyl)ation (MARylation) is reversed by a number of enzymes including a family of MacroD-type macrodomain-containing mono(ADP-ribose) (MAR) hydrolases. Though it has been inferred from various chemical tests that these enzymes have specificity for MARylated aspartate and glutamate residues in vitro, the amino acid and site specificity of different family members are often not unambiguously defined. Here we describe a mass spectrometry-based assay to determine the site specificity of MAR hydrolases in vitro.
Keywords: ADP-ribosylation; ADP-ribosylhydrolase; Macrodomain; Mass spectrometry.
Figures

Similar articles
-
Generating Protein-Linked and Protein-Free Mono-, Oligo-, and Poly(ADP-Ribose) In Vitro.Methods Mol Biol. 2018;1813:91-108. doi: 10.1007/978-1-4939-8588-3_7. Methods Mol Biol. 2018. PMID: 30097863 Free PMC article.
-
Mono-ADP-Ribosylation Catalyzed by Arginine-Specific ADP-Ribosyltransferases.Methods Mol Biol. 2018;1813:149-165. doi: 10.1007/978-1-4939-8588-3_10. Methods Mol Biol. 2018. PMID: 30097866
-
ADP-Ribosylation, a Multifaceted Posttranslational Modification Involved in the Control of Cell Physiology in Health and Disease.Chem Rev. 2018 Feb 14;118(3):1092-1136. doi: 10.1021/acs.chemrev.7b00122. Epub 2017 Nov 27. Chem Rev. 2018. PMID: 29172462 Review.
-
Assessment of Intracellular Auto-Modification Levels of ARTD10 Using Mono-ADP-Ribose-Specific Macrodomains 2 and 3 of Murine Artd8.Methods Mol Biol. 2018;1813:41-63. doi: 10.1007/978-1-4939-8588-3_4. Methods Mol Biol. 2018. PMID: 30097860
-
ADP-ribosylarginine hydrolases and ADP-ribosyltransferases. Partners in ADP-ribosylation cycles.Adv Exp Med Biol. 1997;419:25-33. doi: 10.1007/978-1-4419-8632-0_3. Adv Exp Med Biol. 1997. PMID: 9193633 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

