Mechanistic Studies of Radical SAM Enzymes: Pyruvate Formate-Lyase Activating Enzyme and Lysine 2,3-Aminomutase Case Studies
- PMID: 30097096
- PMCID: PMC8956242
- DOI: 10.1016/bs.mie.2018.04.013
Mechanistic Studies of Radical SAM Enzymes: Pyruvate Formate-Lyase Activating Enzyme and Lysine 2,3-Aminomutase Case Studies
Abstract
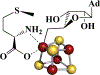
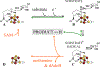
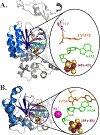
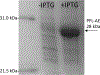
The radical SAM enzyme superfamily is large and diverse, with ever-increasing numbers of examples of characterized reactions. This chapter focuses on the methodology we have developed over the last 25 years for working with these enzymes, with the specific examples discussed being the pyruvate formate-lyase activating enzyme (PFL-AE) and lysine 2,3-aminomutase (LAM). Both enzymes are purified from overexpressing Escherichia coli, but differ in that PFL-AE is expressed without an affinity tag and does not require iron-sulfur cluster reconstitution, while LAM purification is carried out through use of a His6 affinity tag and the enzyme benefits from cluster reconstitution. Because of radical SAM enzymes' catalytic need for a [4Fe-4S] cluster, we present methods for characterization and incorporation of a full [4Fe-4S] cluster in addition to enzyme activity assay protocols. Synthesis of SAM (S-adenosyl-l-methionine) and its analogs have played an important role in our mechanistic studies of radical SAM enzymes, and their synthetic methods are also presented in detail.
Keywords: Glycyl radical enzyme; Lysine 2,3-aminomutase; Purification; Pyruvate formate-lyase; Radical SAM; Reconstitution; S-adenosylmethionine.
© 2018 Elsevier Inc. All rights reserved.
Figures
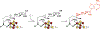

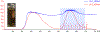
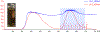
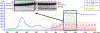
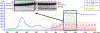



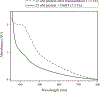
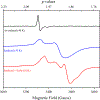


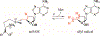



Similar articles
-
Mechanism of Radical Initiation in the Radical S-Adenosyl-l-methionine Superfamily.Acc Chem Res. 2018 Nov 20;51(11):2611-2619. doi: 10.1021/acs.accounts.8b00356. Epub 2018 Oct 15. Acc Chem Res. 2018. PMID: 30346729 Free PMC article. Review.
-
Structural studies of the interaction of S-adenosylmethionine with the [4Fe-4S] clusters in biotin synthase and pyruvate formate-lyase activating enzyme.Protein Sci. 2003 Jul;12(7):1573-7. doi: 10.1110/ps.0302203. Protein Sci. 2003. PMID: 12824504 Free PMC article.
-
Coordination and mechanism of reversible cleavage of S-adenosylmethionine by the [4Fe-4S] center in lysine 2,3-aminomutase.J Am Chem Soc. 2003 Oct 1;125(39):11788-9. doi: 10.1021/ja036120z. J Am Chem Soc. 2003. PMID: 14505379
-
Binding energy in the one-electron reductive cleavage of S-adenosylmethionine in lysine 2,3-aminomutase, a radical SAM enzyme.Biochemistry. 2007 Nov 13;46(45):12889-95. doi: 10.1021/bi701745h. Epub 2007 Oct 18. Biochemistry. 2007. PMID: 17944492 Free PMC article.
-
S-adenosylmethionine as an oxidant: the radical SAM superfamily.Trends Biochem Sci. 2007 Mar;32(3):101-10. doi: 10.1016/j.tibs.2007.01.002. Epub 2007 Feb 8. Trends Biochem Sci. 2007. PMID: 17291766 Review.
Cited by
-
Pyruvate formate-lyase activating enzyme: The catalytically active 5'-deoxyadenosyl radical caught in the act of H-atom abstraction.Proc Natl Acad Sci U S A. 2023 Nov 21;120(47):e2314696120. doi: 10.1073/pnas.2314696120. Epub 2023 Nov 13. Proc Natl Acad Sci U S A. 2023. PMID: 37956301 Free PMC article.
-
Anaerobic benzene oxidation in Geotalea daltonii involves activation by methylation and is regulated by the transition state regulator AbrB.Appl Environ Microbiol. 2024 Oct 23;90(10):e0085624. doi: 10.1128/aem.00856-24. Epub 2024 Sep 17. Appl Environ Microbiol. 2024. PMID: 39287397 Free PMC article.
-
The Elusive 5'-Deoxyadenosyl Radical: Captured and Characterized by Electron Paramagnetic Resonance and Electron Nuclear Double Resonance Spectroscopies.J Am Chem Soc. 2019 Jul 31;141(30):12139-12146. doi: 10.1021/jacs.9b05926. Epub 2019 Jul 22. J Am Chem Soc. 2019. PMID: 31274303 Free PMC article.
-
Radical SAM Enzyme Spore Photoproduct Lyase: Properties of the Ω Organometallic Intermediate and Identification of Stable Protein Radicals Formed during Substrate-Free Turnover.J Am Chem Soc. 2020 Oct 28;142(43):18652-18660. doi: 10.1021/jacs.0c08585. Epub 2020 Oct 15. J Am Chem Soc. 2020. PMID: 32966073 Free PMC article.
-
The Mechanism of Inhibition of Pyruvate Formate Lyase by Methacrylate.J Am Chem Soc. 2023 Oct 18;145(41):22504-22515. doi: 10.1021/jacs.3c07256. Epub 2023 Oct 5. J Am Chem Soc. 2023. PMID: 37797332 Free PMC article.
References
-
- Aasa R, & Vänngård T. (1975). EPR signal intensity and powder shapes: A reexamination. J. Mag. Res, 19, 308–315.
-
- Ashton WT, Brown RD, & Tolman RL (1978). New routes to 1-deoxy-(3,4-dihydro-7,8-dimethyl-2,4-dioxopyrimido[4,5-b]-quinolin-10(2H)yl)-D-ribitols (5-deazariboflavins. J. Heterocyclic. Chem, 15, 489–491.
-
- Ballinger MD, Frey PA, & Reed GH (1992). Structure of a substrate radical intermediate in the reaction of lysine 2,3-aminomutase. Biochemistry, 31(44), 10782–10789. - PubMed
-
- Ballinger MD, Reed GH, & Frey PA (1992). An organic radical in the lysine 2,3-aminomutase reaction. Biochemistry, 31(4), 949–953. - PubMed
-
- Baraniak J, Moss ML, & Frey PA (1989). Lysine 2,3-aminomutase. Support for a mechanism of hydrogen transfer involving S-adenosylmethionine. J. Biol. Chem, 264(3), 1357–1360. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

