Comparative Transcriptomic Analysis of Immune-Related Gene Expression in Duck Embryo Fibroblasts Following Duck Tembusu Virus Infection
- PMID: 30096804
- PMCID: PMC6121397
- DOI: 10.3390/ijms19082328
Comparative Transcriptomic Analysis of Immune-Related Gene Expression in Duck Embryo Fibroblasts Following Duck Tembusu Virus Infection
Abstract
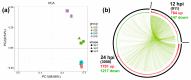
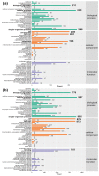
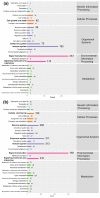
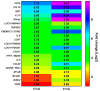
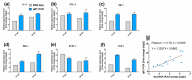
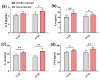
Duck is a major waterfowl species in China, providing high-economic benefit with a population of up to 20⁻30 billion per year. Ducks are commonly affected by severe diseases, including egg-drop syndrome caused by duck Tembusu virus (DTMUV). The immune mechanisms against DTMUV invasion and infection remain poorly understood. In this study, duck embryo fibroblasts (DEFs) were infected with DTMUV and harvested at 12 and 24 h post-infection (hpi), and their genomes were sequenced. In total, 911 (764 upregulated and 147 downregulated genes) and 3008 (1791 upregulated and 1217 downregulated) differentially expressed genes (DEGs) were identified at 12 and 24 hpi, respectively. Kyoto Encyclopedia of Genes and Genomes enrichment analysis revealed that DEGs were considerably enriched in immune-relevant pathways, including Toll-like receptor signaling pathway, Cytosolic DNA-sensing pathway, RIG-I-like receptor signaling pathway, Chemokine signaling pathway, NOD-like receptor signaling pathway, and Hematopoietic cell lineage at both time points. The key DEGs in immune system included those of the cytokines (IFN α2, IL-6, IL-8L, IL-12B, CCR7, CCL19, and CCL20), transcription factors or signaling molecules (IRF7, NF-κB, STAT1, TMEM173, and TNFAIP3), pattern recognition receptors (RIG-I and MDA5), and antigen-presenting proteins (CD44 and CD70). This suggests DTMUV infection induces strong proinflammatory/antiviral effects with enormous production of cytokines. However, these cytokines could not protect DEFs against viral attack. Our data revealed valuable transcriptional information regarding DTMUV-infected DEFs, thereby broadening our understanding of the immune response against DTMUV infection; this information might contribute in developing strategies for controlling the prevalence of DTMUV infection.
Keywords: DTMUV; RNA-seq; duck embryo fibroblast; transcriptome; virus infection.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


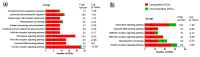

Similar articles
-
Innate immune responses to duck Tembusu virus infection.Vet Res. 2020 Jul 8;51(1):87. doi: 10.1186/s13567-020-00814-9. Vet Res. 2020. PMID: 32641107 Free PMC article. Review.
-
Differently Expression Analysis and Function Prediction of Long Non-coding RNAs in Duck Embryo Fibroblast Cells Infected by Duck Tembusu Virus.Front Immunol. 2020 Aug 4;11:1729. doi: 10.3389/fimmu.2020.01729. eCollection 2020. Front Immunol. 2020. PMID: 32849615 Free PMC article.
-
Duck Tembusu Virus Infection Promotes the Expression of Duck Interferon-Induced Protein 35 to Counteract RIG-I Antiviral Signaling in Duck Embryo Fibroblasts.Front Immunol. 2021 Jul 15;12:711517. doi: 10.3389/fimmu.2021.711517. eCollection 2021. Front Immunol. 2021. PMID: 34335626 Free PMC article.
-
In vivo cellular and molecular study on duck spleen infected by duck Tembusu virus.Vet Microbiol. 2019 Mar;230:32-44. doi: 10.1016/j.vetmic.2018.12.003. Epub 2018 Dec 23. Vet Microbiol. 2019. PMID: 30827402
-
Advancements in Research on Duck Tembusu Virus Infections.Viruses. 2024 May 20;16(5):811. doi: 10.3390/v16050811. Viruses. 2024. PMID: 38793692 Free PMC article. Review.
Cited by
-
DEF Cell-Derived Exosomal miR-148a-5p Promotes DTMUV Replication by Negative Regulating TLR3 Expression.Viruses. 2020 Jan 14;12(1):94. doi: 10.3390/v12010094. Viruses. 2020. PMID: 31947624 Free PMC article.
-
Interferon Regulatory Factors IRF1 and IRF7 Directly Regulate Gene Expression in Bats in Response to Viral Infection.Cell Rep. 2020 Nov 3;33(5):108345. doi: 10.1016/j.celrep.2020.108345. Cell Rep. 2020. PMID: 33147460 Free PMC article.
-
Avian IRF1 and IRF7 Play Overlapping and Distinct Roles in Regulating IFN-Dependent and -Independent Antiviral Responses to Duck Tembusu Virus Infection.Viruses. 2022 Jul 9;14(7):1506. doi: 10.3390/v14071506. Viruses. 2022. PMID: 35891486 Free PMC article.
-
Evolution of Tembusu Virus in Ducks, Chickens, Geese, Sparrows, and Mosquitoes in Northern China.Viruses. 2018 Sep 10;10(9):485. doi: 10.3390/v10090485. Viruses. 2018. PMID: 30201873 Free PMC article.
-
Innate immune responses to duck Tembusu virus infection.Vet Res. 2020 Jul 8;51(1):87. doi: 10.1186/s13567-020-00814-9. Vet Res. 2020. PMID: 32641107 Free PMC article. Review.
References
-
- Fu G., Chen C., Huang Y., Cheng L., Fu Q., Wan C., Shi S., Chen H., Liu W. Comparative analysis of transcriptional profiles of retinoic-acid-induced gene I-like receptors and interferons in seven tissues from ducks infected with avian Tembusu virus. Arch. Virol. 2016;161:11–18. doi: 10.1007/s00705-015-2621-x. - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

