Interphase human chromosome exhibits out of equilibrium glassy dynamics
- PMID: 30089831
- PMCID: PMC6082855
- DOI: 10.1038/s41467-018-05606-6
Interphase human chromosome exhibits out of equilibrium glassy dynamics
Abstract
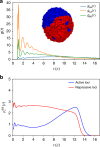
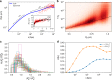
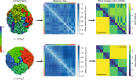
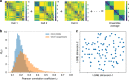
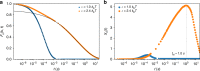
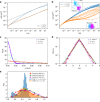
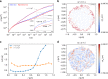
Fingerprints of the three-dimensional organization of genomes have emerged using advances in Hi-C and imaging techniques. However, genome dynamics is poorly understood. Here, we create the chromosome copolymer model (CCM) by representing chromosomes as a copolymer with two epigenetic loci types corresponding to euchromatin and heterochromatin. Using novel clustering techniques, we establish quantitatively that the simulated contact maps and topologically associating domains (TADs) for chromosomes 5 and 10 and those inferred from Hi-C experiments are in good agreement. Chromatin exhibits glassy dynamics with coherent motion on micron scale. The broad distribution of the diffusion exponents of the individual loci, which quantitatively agrees with experiments, is suggestive of highly heterogeneous dynamics. This is reflected in the cell-to-cell variations in the contact maps. Chromosome organization is hierarchical, involving the formation of chromosome droplets (CDs) on genomic scale, coinciding with the TAD size, followed by coalescence of the CDs, reminiscent of Ostwald ripening.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Chain organization of human interphase chromosome determines the spatiotemporal dynamics of chromatin loci.PLoS Comput Biol. 2018 Dec 3;14(12):e1006617. doi: 10.1371/journal.pcbi.1006617. eCollection 2018 Dec. PLoS Comput Biol. 2018. PMID: 30507936 Free PMC article.
-
Simulation of different three-dimensional polymer models of interphase chromosomes compared to experiments-an evaluation and review framework of the 3D genome organization.Semin Cell Dev Biol. 2019 Jun;90:19-42. doi: 10.1016/j.semcdb.2018.07.012. Epub 2018 Aug 24. Semin Cell Dev Biol. 2019. PMID: 30125668 Review.
-
Structural basis for the preservation of a subset of topologically associating domains in interphase chromosomes upon cohesin depletion.Elife. 2024 Mar 19;12:RP88564. doi: 10.7554/eLife.88564. Elife. 2024. PMID: 38502563 Free PMC article.
-
Conformational heterogeneity in human interphase chromosome organization reconciles the FISH and Hi-C paradox.Nat Commun. 2019 Aug 29;10(1):3894. doi: 10.1038/s41467-019-11897-0. Nat Commun. 2019. PMID: 31467267 Free PMC article.
-
Structural-functional model of the mitotic chromosome.Biochemistry (Mosc). 2006 Jan;71(1):1-9. doi: 10.1134/s0006297906010019. Biochemistry (Mosc). 2006. PMID: 16457612 Review.
Cited by
-
Characterizing the variation in chromosome structure ensembles in the context of the nuclear microenvironment.PLoS Comput Biol. 2022 Aug 15;18(8):e1010392. doi: 10.1371/journal.pcbi.1010392. eCollection 2022 Aug. PLoS Comput Biol. 2022. PMID: 35969616 Free PMC article.
-
Large-scale simulations of nucleoprotein complexes: ribosomes, nucleosomes, chromatin, chromosomes and CRISPR.Curr Opin Struct Biol. 2019 Apr;55:104-113. doi: 10.1016/j.sbi.2019.03.004. Epub 2019 May 21. Curr Opin Struct Biol. 2019. PMID: 31125796 Free PMC article. Review.
-
Heterogeneous interactions and polymer entropy decide organization and dynamics of chromatin domains.Biophys J. 2022 Jul 19;121(14):2794-2812. doi: 10.1016/j.bpj.2022.06.008. Epub 2022 Jun 6. Biophys J. 2022. PMID: 35672951 Free PMC article.
-
A Liquid State Perspective on Dynamics of Chromatin Compartments.Front Mol Biosci. 2022 Jan 13;8:781981. doi: 10.3389/fmolb.2021.781981. eCollection 2021. Front Mol Biosci. 2022. PMID: 35096966 Free PMC article. Review.
-
Leveraging polymer modeling to reconstruct chromatin connectivity from live images.Biophys J. 2023 Sep 5;122(17):3532-3540. doi: 10.1016/j.bpj.2023.08.001. Epub 2023 Aug 4. Biophys J. 2023. PMID: 37542372 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

