Making glycoproteins a little bit sweeter with PDB-REDO
- PMID: 30084395
- PMCID: PMC6096482
- DOI: 10.1107/S2053230X18004016
Making glycoproteins a little bit sweeter with PDB-REDO
Abstract
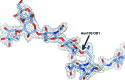
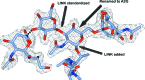
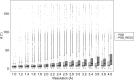
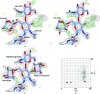
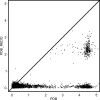
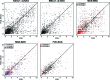
Glycosylation is one of the most common forms of protein post-translational modification, but is also the most complex. Dealing with glycoproteins in structure model building, refinement, validation and PDB deposition is more error-prone than dealing with nonglycosylated proteins owing to limitations of the experimental data and available software tools. Also, experimentalists are typically less experienced in dealing with carbohydrate residues than with amino-acid residues. The results of the reannotation and re-refinement by PDB-REDO of 8114 glycoprotein structure models from the Protein Data Bank are analyzed. The positive aspects of 3620 reannotations and subsequent refinement, as well as the remaining challenges to obtaining consistently high-quality carbohydrate models, are discussed.
Keywords: PDB-REDO; carbohydrates; glycoproteins; pdb-care; validation.
open access.
Figures

Similar articles
-
Building and rebuilding N-glycans in protein structure models.Acta Crystallogr D Struct Biol. 2019 Apr 1;75(Pt 4):416-425. doi: 10.1107/S2059798319003875. Epub 2019 Apr 4. Acta Crystallogr D Struct Biol. 2019. PMID: 30988258 Free PMC article.
-
Carbohydrate Structure Suite (CSS): analysis of carbohydrate 3D structures derived from the PDB.Nucleic Acids Res. 2005 Jan 1;33(Database issue):D242-6. doi: 10.1093/nar/gki013. Nucleic Acids Res. 2005. PMID: 15608187 Free PMC article.
-
PDB_REDO: constructive validation, more than just looking for errors.Acta Crystallogr D Biol Crystallogr. 2012 Apr;68(Pt 4):484-96. doi: 10.1107/S0907444911054515. Epub 2012 Mar 16. Acta Crystallogr D Biol Crystallogr. 2012. PMID: 22505269 Free PMC article.
-
Data mining the PDB for glyco-related data.Methods Mol Biol. 2009;534:293-310. doi: 10.1007/978-1-59745-022-5_21. Methods Mol Biol. 2009. PMID: 19277543 Review.
-
New Biological Insights from Better Structure Models.J Mol Biol. 2016 Mar 27;428(6):1375-1393. doi: 10.1016/j.jmb.2016.02.002. Epub 2016 Feb 8. J Mol Biol. 2016. PMID: 26869101 Review.
Cited by
-
Three-Dimensional Structures of Carbohydrates and Where to Find Them.Int J Mol Sci. 2020 Oct 18;21(20):7702. doi: 10.3390/ijms21207702. Int J Mol Sci. 2020. PMID: 33081008 Free PMC article. Review.
-
Building and rebuilding N-glycans in protein structure models.Acta Crystallogr D Struct Biol. 2019 Apr 1;75(Pt 4):416-425. doi: 10.1107/S2059798319003875. Epub 2019 Apr 4. Acta Crystallogr D Struct Biol. 2019. PMID: 30988258 Free PMC article.
-
Modelling covalent linkages in CCP4.Acta Crystallogr D Struct Biol. 2021 Jun 1;77(Pt 6):712-726. doi: 10.1107/S2059798321001753. Epub 2021 May 19. Acta Crystallogr D Struct Biol. 2021. PMID: 34076587 Free PMC article.
-
The human gut symbiont Ruminococcus gnavus shows specificity to blood group A antigen during mucin glycan foraging: Implication for niche colonisation in the gastrointestinal tract.PLoS Biol. 2021 Dec 22;19(12):e3001498. doi: 10.1371/journal.pbio.3001498. eCollection 2021 Dec. PLoS Biol. 2021. PMID: 34936658 Free PMC article.
-
Monitoring carbohydrate 3D structure quality with the Privateer database.Beilstein J Org Chem. 2024 Apr 24;20:931-939. doi: 10.3762/bjoc.20.83. eCollection 2024. Beilstein J Org Chem. 2024. PMID: 38711584 Free PMC article.
References
-
- Agirre, J., Davies, G., Wilson, K. & Cowtan, K. (2015). Nature Chem. Biol. 11, 303. - PubMed
-
- Agirre, J., Davies, G. J., Wilson, K. S. & Cowtan, K. D. (2017). Curr. Opin. Struct. Biol. 44, 39–47. - PubMed
-
- Agirre, J., Iglesias-Fernández, J., Rovira, C., Davies, G. J., Wilson, K. S. & Cowtan, K. D. (2015). Nature Struct. Mol. Biol. 22, 833–834. - PubMed
-
- Bernstein, F. C., Koetzle, T. F., Williams, G. J., Meyer, E. F., Brice, M. D., Rodgers, J. R., Kennard, O., Shimanouchi, T. & Tasumi, M. (1977). J. Mol. Biol. 112, 535–542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

