A Virus in American Blackcurrant (Ribes americanum) with Distinct Genome Features Reshapes Classification in the Tymovirales
- PMID: 30081487
- PMCID: PMC6115964
- DOI: 10.3390/v10080406
A Virus in American Blackcurrant (Ribes americanum) with Distinct Genome Features Reshapes Classification in the Tymovirales
Abstract
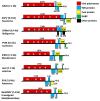
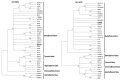
A novel virus with distinct genome features was discovered by high throughput sequencing in a symptomatic blackcurrant plant. The virus, tentatively named Ribes americanum virus A (RAVA), has distinct genome organization and molecular features bridging genera in the order Tymovirales. The genome consists of 7106 nucleotides excluding the poly(A) tail. Five open reading frames were identified, with the first encoding a putative viral replicase with methyl transferase (MTR), AlkB, helicase, and RNA dependent RNA polymerase (RdRp) domains. The genome organization downstream of the replicase resembles that of members of the order Tymovirales with an unconventional triple gene block (TGB) movement protein arrangement with none of the other four putative proteins exhibiting significant homology to viral proteins. Phylogenetic analysis using replicase conserved motifs loosely placed RAVA within the Betaflexiviridae. Data strongly suggest that RAVA is a novel virus that should be classified as a species in a new genus in the Betaflexiviridae or a new family within the order Tymovirales.
Keywords: Betaflexiviridae; Ribes americanum virus A; blackcurrant; characterization; detection.
Conflict of interest statement
The authors declare no conflict of interest. The funding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures

Similar articles
-
Molecular characterization of a novel capillovirus from red currant.Arch Virol. 2016 Apr;161(4):1083-6. doi: 10.1007/s00705-016-2752-8. Epub 2016 Jan 11. Arch Virol. 2016. PMID: 26754736
-
Molecular characterization of a new vitivirus discovered in a blueberry plant with green mosaic symptoms.Arch Virol. 2019 Oct;164(10):2609-2611. doi: 10.1007/s00705-019-04344-4. Epub 2019 Jul 16. Arch Virol. 2019. PMID: 31312966
-
Molecular characterization of a novel rhabdovirus infecting blackcurrant identified by high-throughput sequencing.Arch Virol. 2018 May;163(5):1363-1366. doi: 10.1007/s00705-018-3709-x. Epub 2018 Jan 24. Arch Virol. 2018. PMID: 29368064
-
Family Flexiviridae: a case study in virion and genome plasticity.Annu Rev Phytopathol. 2007;45:73-100. doi: 10.1146/annurev.phyto.45.062806.094401. Annu Rev Phytopathol. 2007. PMID: 17362202 Review.
-
Status of the current vitivirus taxonomy.Arch Virol. 2020 Feb;165(2):451-458. doi: 10.1007/s00705-019-04500-w. Epub 2019 Dec 16. Arch Virol. 2020. PMID: 31845154 Review.
Cited by
-
Pest categorisation of non-EU viruses of Ribes L.EFSA J. 2019 Nov 15;17(11):e05859. doi: 10.2903/j.efsa.2019.5859. eCollection 2019 Nov. EFSA J. 2019. PMID: 32626160 Free PMC article.
-
Small hydrophobic viral proteins involved in intercellular movement of diverse plant virus genomes.AIMS Microbiol. 2020 Sep 21;6(3):305-329. doi: 10.3934/microbiol.2020019. eCollection 2020. AIMS Microbiol. 2020. PMID: 33134746 Free PMC article. Review.
References
-
- Postman J., Hummer K., Stover E., Krueger R., Forsline P., Grauke L.J., Zee F., Ayala-Silva T., Irish B. Fruit and nut genebanks in the US national plant germplasm system. HortScience. 2006;41:1188–1194.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

