Porcine Adenovirus Type 3 E3 Encodes a Structural Protein Essential for Capsid Stability and Production of Infectious Progeny Virions
- PMID: 30068639
- PMCID: PMC6158433
- DOI: 10.1128/JVI.00680-18
Porcine Adenovirus Type 3 E3 Encodes a Structural Protein Essential for Capsid Stability and Production of Infectious Progeny Virions
Abstract
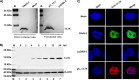
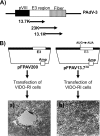
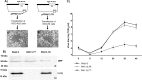
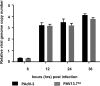
The adenovirus E3 region encodes proteins that are not essential for viral replication in vitro The porcine adenovirus type 3 (PAdV-3) E3 region encodes three proteins, including 13.7K. Here, we report that 13.7K is expressed as an early protein, which localizes to the nucleus of infected cells. The 13.7K protein is a structural protein, as it is incorporated in CsCl-purified virions. The 13.7K protein appears to be essential for PAdV-3 replication, as mutant PAV13.73A expressing a mutated 13.7K could be isolated only in VIDO AS2 cells expressing the 13.7K protein. Analysis of PAV13.73A suggested that even in the presence of reduced levels of some late viral proteins, there appeared to be no effect on virus assembly and production of mature virions. Further analysis of CsCl-purified PAV13.73A by transmission electron microscopy revealed the presence of disrupted/broken capsids, suggesting that inactivation of 13.7K protein expression may produce fragile capsids. Our results suggest that the PAdV-3 E3 region-encoded 13.7K protein is a capsid protein, which appears to be essential for the formation of stable capsids and production of infectious progeny virions.IMPORTANCE Although E3 region-encoded proteins are involved in the modulation of leukocyte functions (N. Arnberg, Proc Natl Acad Sci U S A 110:19976-19977, 2013) and inducing a lytic infection of lymphocytes (V. K. Murali, D. A. Ornelles, L. R. Gooding, H. T. Wilms, W. Huang, A. E. Tollefson, W. S. Wold, and C. Garnett-Benson, J Virol 88:903-912, 2014), none of the E3 proteins appear to be a component of virion capsid or required for replication of adenovirus. Here, we demonstrate that the 13.7K protein encoded by the E3 region of porcine adenovirus type 3 is a component of progeny virion capsids and appears to be essential for maintaining the integrity of virion capsid and production of infectious progeny virions. To our knowledge, this is the first report to suggest that an adenovirus E3-encoded protein is an essential structural protein.
Keywords: 13.7K; E3; PAdV-3; essential protein; porcine adenovirus; structural protein.
Copyright © 2018 American Society for Microbiology.
Figures





Similar articles
-
The 10,400- and 14,500-dalton proteins encoded by region E3 of adenovirus function together to protect many but not all mouse cell lines against lysis by tumor necrosis factor.J Virol. 1991 Aug;65(8):4114-23. doi: 10.1128/JVI.65.8.4114-4123.1991. J Virol. 1991. PMID: 1830111 Free PMC article.
-
A Single Maturation Cleavage Site in Adenovirus Impacts Cell Entry and Capsid Assembly.J Virol. 2015 Oct 21;90(1):521-32. doi: 10.1128/JVI.02014-15. Print 2016 Jan 1. J Virol. 2015. PMID: 26491163 Free PMC article.
-
Adenovirus E3 14.7K protein functions in the absence of other adenovirus proteins to protect transfected cells from tumor necrosis factor cytolysis.J Virol. 1991 May;65(5):2629-39. doi: 10.1128/JVI.65.5.2629-2639.1991. J Virol. 1991. PMID: 1850036 Free PMC article.
-
Adenovirus region E3 proteins that prevent cytolysis by cytotoxic T cells and tumor necrosis factor.Mol Biol Med. 1989 Oct;6(5):433-52. Mol Biol Med. 1989. PMID: 2533958 Review.
-
Helical capsids of plant viruses: architecture with structural lability.J Gen Virol. 2016 Aug;97(8):1739-1754. doi: 10.1099/jgv.0.000524. Epub 2016 Jun 15. J Gen Virol. 2016. PMID: 27312096 Review.
Cited by
-
Deletion of the H240R Gene of African Swine Fever Virus Decreases Infectious Progeny Virus Production Due to Aberrant Virion Morphogenesis and Enhances Inflammatory Cytokine Expression in Porcine Macrophages.J Virol. 2022 Feb 9;96(3):e0166721. doi: 10.1128/JVI.01667-21. Epub 2021 Nov 17. J Virol. 2022. PMID: 34787458 Free PMC article.
References
-
- Kadoi K, Iwabuchi M, Satoh T, Katase T, Kawaji T, Morichi T. 1997. Adenovirus isolation from spleen lymphocytes of apparently healthy pigs. New Microbiol 20:215–220. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

