Species-Specific Changes in a Primate Transcription Factor Network Provide Insights into the Molecular Evolution of the Primate Prefrontal Cortex
- PMID: 30059966
- PMCID: PMC6105097
- DOI: 10.1093/gbe/evy149
Species-Specific Changes in a Primate Transcription Factor Network Provide Insights into the Molecular Evolution of the Primate Prefrontal Cortex
Abstract
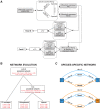
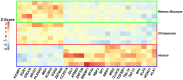
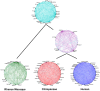
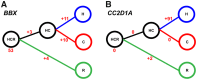
The human prefrontal cortex (PFC) differs from that of other primates with respect to size, histology, and functional abilities. Here, we analyzed genome-wide expression data of humans, chimpanzees, and rhesus macaques to discover evolutionary changes in transcription factor (TF) networks that may underlie these phenotypic differences. We determined the co-expression networks of all TFs with species-specific expression including their potential target genes and interaction partners in the PFC of all three species. Integrating these networks allowed us inferring an ancestral network for all three species. This ancestral network as well as the networks for each species is enriched for genes involved in forebrain development, axonogenesis, and synaptic transmission. Our analysis allows us to directly compare the networks of each species to determine which links have been gained or lost during evolution. Interestingly, we detected that most links were gained on the human lineage, indicating increase TF cooperativity in humans. By comparing network changes between different tissues, we discovered that in brain tissues, but not in the other tissues, the human networks always had the highest connectivity. To pinpoint molecular changes underlying species-specific phenotypes, we analyzed the sub-networks of TFs derived only from genes with species-specific expression changes in the PFC. These sub-networks differed significantly in structure and function between the human and chimpanzee. For example, the human-specific sub-network is enriched for TFs implicated in cognitive disorders and for genes involved in synaptic plasticity and cognitive functions. Our results suggest evolutionary changes in TF networks that might have shaped morphological and functional differences between primate brains, in particular in the human PFC.
Figures

Similar articles
-
Differences in human and chimpanzee gene expression patterns define an evolving network of transcription factors in brain.Proc Natl Acad Sci U S A. 2009 Dec 29;106(52):22358-63. doi: 10.1073/pnas.0911376106. Epub 2009 Dec 10. Proc Natl Acad Sci U S A. 2009. PMID: 20007773 Free PMC article.
-
Evolutionary and ontogenetic changes in RNA editing in human, chimpanzee, and macaque brains.RNA. 2013 Dec;19(12):1693-702. doi: 10.1261/rna.039206.113. Epub 2013 Oct 23. RNA. 2013. PMID: 24152549 Free PMC article.
-
Epigenomic annotation of gene regulatory alterations during evolution of the primate brain.Nat Neurosci. 2016 Mar;19(3):494-503. doi: 10.1038/nn.4229. Epub 2016 Jan 25. Nat Neurosci. 2016. PMID: 26807951
-
Comparative primate genomics: the year of the chimpanzee.Curr Opin Genet Dev. 2004 Dec;14(6):650-6. doi: 10.1016/j.gde.2004.08.007. Curr Opin Genet Dev. 2004. PMID: 15531160 Review.
-
Plasticity of the primate prefrontal cortex.Neuroscientist. 2007 Jun;13(3):229-40. doi: 10.1177/1073858406298554. Neuroscientist. 2007. PMID: 17519366 Review.
Cited by
-
Genomics at cellular resolution: insights into cognitive disorders and their evolution.Hum Mol Genet. 2020 Sep 30;29(R1):R1-R9. doi: 10.1093/hmg/ddaa117. Hum Mol Genet. 2020. PMID: 32566943 Free PMC article. Review.
-
Identifying nootropic drug targets via large-scale cognitive GWAS and transcriptomics.Neuropsychopharmacology. 2021 Sep;46(10):1788-1801. doi: 10.1038/s41386-021-01023-4. Epub 2021 May 25. Neuropsychopharmacology. 2021. PMID: 34035472 Free PMC article.
-
Accelerated evolution of oligodendrocytes in the human brain.Proc Natl Acad Sci U S A. 2019 Nov 26;116(48):24334-24342. doi: 10.1073/pnas.1907982116. Epub 2019 Nov 11. Proc Natl Acad Sci U S A. 2019. PMID: 31712436 Free PMC article.
-
Dendritic Spines: Synaptogenesis and Synaptic Pruning for the Developmental Organization of Brain Circuits.Adv Neurobiol. 2023;34:143-221. doi: 10.1007/978-3-031-36159-3_4. Adv Neurobiol. 2023. PMID: 37962796
-
A strategy for designing allosteric modulators of transcription factor dimerization.Proc Natl Acad Sci U S A. 2020 Feb 4;117(5):2683-2686. doi: 10.1073/pnas.1915531117. Epub 2020 Jan 17. Proc Natl Acad Sci U S A. 2020. PMID: 31953259 Free PMC article.
References
-
- Allen NC, et al. 2008. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 40(7):827–834. - PubMed
-
- Anders S, Huber W.. 2012. Differential expression of RNA-Seq data at the gene level–the DESeq package. Heidelberg (Germany): European Molecular Biology Laboratory (EMBL).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

