Metagenomic sequencing suggests a diversity of RNA interference-like responses to viruses across multicellular eukaryotes
- PMID: 30059538
- PMCID: PMC6085071
- DOI: 10.1371/journal.pgen.1007533
Metagenomic sequencing suggests a diversity of RNA interference-like responses to viruses across multicellular eukaryotes
Erratum in
-
Correction: Metagenomic sequencing suggests a diversity of RNA interference-like responses to viruses across multicellular eukaryotes.PLoS Genet. 2020 Mar 2;16(3):e1008679. doi: 10.1371/journal.pgen.1008679. eCollection 2020 Mar. PLoS Genet. 2020. PMID: 32119721 Free PMC article.
Abstract
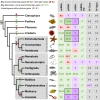
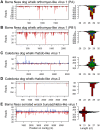
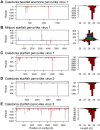
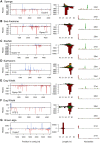
RNA interference (RNAi)-related pathways target viruses and transposable element (TE) transcripts in plants, fungi, and ecdysozoans (nematodes and arthropods), giving protection against infection and transmission. In each case, this produces abundant TE and virus-derived 20-30nt small RNAs, which provide a characteristic signature of RNAi-mediated defence. The broad phylogenetic distribution of the Argonaute and Dicer-family genes that mediate these pathways suggests that defensive RNAi is ancient, and probably shared by most animal (metazoan) phyla. Indeed, while vertebrates had been thought an exception, it has recently been argued that mammals also possess an antiviral RNAi pathway, although its immunological relevance is currently uncertain and the viral small RNAs (viRNAs) are not easily detectable. Here we use a metagenomic approach to test for the presence of viRNAs in five species from divergent animal phyla (Porifera, Cnidaria, Echinodermata, Mollusca, and Annelida), and in a brown alga-which represents an independent origin of multicellularity from plants, fungi, and animals. We use metagenomic RNA sequencing to identify around 80 virus-like contigs in these lineages, and small RNA sequencing to identify viRNAs derived from those viruses. We identified 21U small RNAs derived from an RNA virus in the brown alga, reminiscent of plant and fungal viRNAs, despite the deep divergence between these lineages. However, contrary to our expectations, we were unable to identify canonical (i.e. Drosophila- or nematode-like) viRNAs in any of the animals, despite the widespread presence of abundant micro-RNAs, and somatic transposon-derived piwi-interacting RNAs. We did identify a distinctive group of small RNAs derived from RNA viruses in the mollusc. However, unlike ecdysozoan viRNAs, these had a piRNA-like length distribution but lacked key signatures of piRNA biogenesis. We also identified primary piRNAs derived from putatively endogenous copies of DNA viruses in the cnidarian and the echinoderm, and an endogenous RNA virus in the mollusc. The absence of canonical virus-derived small RNAs from our samples may suggest that the majority of animal phyla lack an antiviral RNAi response. Alternatively, these phyla could possess an antiviral RNAi response resembling that reported for vertebrates, with cryptic viRNAs not detectable through simple metagenomic sequencing of wild-type individuals. In either case, our findings show that the antiviral RNAi responses of arthropods and nematodes, which are highly divergent from each other and from that of plants and fungi, are also highly diverged from the most likely ancestral metazoan state.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
RNA-based viral immunity initiated by the Dicer family of host immune receptors.Immunol Rev. 2009 Jan;227(1):176-88. doi: 10.1111/j.1600-065X.2008.00722.x. Immunol Rev. 2009. PMID: 19120484 Free PMC article. Review.
-
Endogenous Viral Element-Derived Piwi-Interacting RNAs (piRNAs) Are Not Required for Production of Ping-Pong-Dependent piRNAs from Diaphorina citri Densovirus.mBio. 2020 Sep 29;11(5):e02209-20. doi: 10.1128/mBio.02209-20. mBio. 2020. PMID: 32994324 Free PMC article.
-
Caenorhabditis elegans RIG-I Homolog Mediates Antiviral RNA Interference Downstream of Dicer-Dependent Biogenesis of Viral Small Interfering RNAs.mBio. 2017 Mar 21;8(2):e00264-17. doi: 10.1128/mBio.00264-17. mBio. 2017. PMID: 28325765 Free PMC article.
-
Mechanism and Function of Antiviral RNA Interference in Mice.mBio. 2020 Aug 4;11(4):e03278-19. doi: 10.1128/mBio.03278-19. mBio. 2020. PMID: 32753500 Free PMC article.
-
PIWI pathway against viruses in insects.Wiley Interdiscip Rev RNA. 2019 Nov;10(6):e1555. doi: 10.1002/wrna.1555. Epub 2019 Jun 10. Wiley Interdiscip Rev RNA. 2019. PMID: 31183996 Review.
Cited by
-
The origin of RNA interference: Adaptive or neutral evolution?PLoS Biol. 2022 Jun 29;20(6):e3001715. doi: 10.1371/journal.pbio.3001715. eCollection 2022 Jun. PLoS Biol. 2022. PMID: 35767561 Free PMC article.
-
Metatranscriptomic Identification of Diverse and Divergent RNA Viruses in Green and Chlorarachniophyte Algae Cultures.Viruses. 2020 Oct 19;12(10):1180. doi: 10.3390/v12101180. Viruses. 2020. PMID: 33086653 Free PMC article.
-
Metagenomic Assessment of DNA Viral Diversity in Freshwater Sponges, Baikalospongia bacillifera.Microorganisms. 2022 Feb 21;10(2):480. doi: 10.3390/microorganisms10020480. Microorganisms. 2022. PMID: 35208935 Free PMC article.
-
Initial Virome Characterization of the Common Cnidarian Lab Model Nematostella vectensis.Viruses. 2020 Feb 15;12(2):218. doi: 10.3390/v12020218. Viruses. 2020. PMID: 32075325 Free PMC article.
-
Functional Characterization of the Cnidarian Antiviral Immune Response Reveals Ancestral Complexity.Mol Biol Evol. 2021 Sep 27;38(10):4546-4561. doi: 10.1093/molbev/msab197. Mol Biol Evol. 2021. PMID: 34180999 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

