Mapping of Plasma Membrane Proteins Interacting With Arabidopsis thaliana Flotillin 2
- PMID: 30050548
- PMCID: PMC6052134
- DOI: 10.3389/fpls.2018.00991
Mapping of Plasma Membrane Proteins Interacting With Arabidopsis thaliana Flotillin 2
Abstract
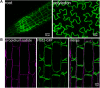
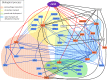
Arabidopsis flotillin 2 (At5g25260) belongs to the group of plant flotillins, which are not well characterized. In contrast, metazoan flotillins are well known as plasma membrane proteins associated with membrane microdomains that act as a signaling hub. The similarity of plant and metazoan flotillins, whose functions most likely consist of affecting other proteins via protein-protein interactions, determines the necessity of detecting their interacting partners in plants. Nevertheless, identifying the proteins that form complexes on the plasma membrane is a challenging task due to their low abundance and hydrophobic character. Here we present an approach for mapping Arabidopsis thaliana flotillin 2 plasma membrane interactors, based on the immunoaffinity purification of crosslinked and enriched plasma membrane proteins with mass spectrometry detection. Using this approach, 61 proteins were enriched in the AtFlot-GFP plasma membrane fraction, and 19 of them were proposed to be flotillin 2 interaction partners. Among our proposed partners of Flot2, proteins playing a role in the plant response to various biotic and abiotic stresses were detected. Additionally, the use of the split-ubiquitin yeast system helped us to confirm that plasma-membrane ATPase 1, early-responsive to dehydration stress protein 4, syntaxin-71, harpin-induced protein-like 3, hypersensitive-induced response protein 2 and two aquaporin isoforms interact with flotillin 2 directly. Based on the results of our study and the reported properties of Flot2 interactors, we propose that Flot2 complexes may be involved in plant-pathogen interactions, water transport and intracellular trafficking.
Keywords: Arabidopsis flotillin 2; immunopurification; intracellular trafficking; mass spectrometry; plant–pathogen interaction; protein–protein interactions; split-ubiquitin yeast system; water transport.
Figures



Similar articles
-
Cell wall contributes to the stability of plasma membrane nanodomain organization of Arabidopsis thaliana FLOTILLIN2 and HYPERSENSITIVE INDUCED REACTION1 proteins.Plant J. 2020 Feb;101(3):619-636. doi: 10.1111/tpj.14566. Epub 2019 Nov 11. Plant J. 2020. PMID: 31610051
-
Role of dynamin and clathrin in the cellular trafficking of flotillins.FEBS J. 2014 Jul;281(13):2956-76. doi: 10.1111/febs.12834. Epub 2014 May 27. FEBS J. 2014. PMID: 24809731
-
Flotillins: At the Intersection of Protein S-Palmitoylation and Lipid-Mediated Signaling.Int J Mol Sci. 2020 Mar 26;21(7):2283. doi: 10.3390/ijms21072283. Int J Mol Sci. 2020. PMID: 32225034 Free PMC article. Review.
-
Flotillin membrane domains in cancer.Cancer Metastasis Rev. 2020 Jun;39(2):361-374. doi: 10.1007/s10555-020-09873-y. Cancer Metastasis Rev. 2020. PMID: 32297092 Free PMC article. Review.
-
Environmental Cues Contribute to Dynamic Plasma Membrane Organization of Nanodomains Containing Flotillin-1 and Hypersensitive Induced Reaction-1 Proteins in Arabidopsis thaliana.Front Plant Sci. 2022 May 10;13:897594. doi: 10.3389/fpls.2022.897594. eCollection 2022. Front Plant Sci. 2022. PMID: 35620697 Free PMC article.
Cited by
-
Involvement of the Membrane Nanodomain Protein, AtFlot1, in Vesicular Transport of Plasma Membrane H+-ATPase in Arabidopsis thaliana under Salt Stress.Int J Mol Sci. 2023 Jan 8;24(2):1251. doi: 10.3390/ijms24021251. Int J Mol Sci. 2023. PMID: 36674767 Free PMC article.
-
Membrane nanodomains and transport functions in plant.Plant Physiol. 2021 Dec 4;187(4):1839-1855. doi: 10.1093/plphys/kiab312. Plant Physiol. 2021. PMID: 35235669 Free PMC article. Review.
-
The Arabidopsis Proteins AtNHR2A and AtNHR2B Are Multi-Functional Proteins Integrating Plant Immunity With Other Biological Processes.Front Plant Sci. 2020 Mar 4;11:232. doi: 10.3389/fpls.2020.00232. eCollection 2020. Front Plant Sci. 2020. PMID: 32194606 Free PMC article.
-
An Integrated Approach for the Efficient Extraction and Solubilization of Rice Microsomal Membrane Proteins for High-Throughput Proteomics.Front Plant Sci. 2021 Sep 9;12:723369. doi: 10.3389/fpls.2021.723369. eCollection 2021. Front Plant Sci. 2021. PMID: 34567038 Free PMC article.
-
Dynamics and Endocytosis of Flot1 in Arabidopsis Require CPI1 Function.Int J Mol Sci. 2020 Feb 25;21(5):1552. doi: 10.3390/ijms21051552. Int J Mol Sci. 2020. PMID: 32106431 Free PMC article.
References
-
- Ascencio-Ibanez J. T., Sozzani R., Lee T. J., Chu T. M., Wolfinger R. D., Cella R., et al. (2008). Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148 436–454. 10.1104/pp.108.121038 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

