The mutational landscape of recurrent versus nonrecurrent human papillomavirus-related oropharyngeal cancer
- PMID: 30046007
- PMCID: PMC6124437
- DOI: 10.1172/jci.insight.99327
The mutational landscape of recurrent versus nonrecurrent human papillomavirus-related oropharyngeal cancer
Abstract
Background: Human papillomavirus-related (HPV-related) oropharyngeal squamous cell carcinomas (OPSCCs) have an excellent response rate to platinum-based chemoradiotherapy. Genomic differences between primary HPV-related OPSCCs that do or do not recur are unknown. Furthermore, it is unclear if HPV-related OPSCCs that recur share a genomic landscape with HPV-negative head and neck cancers (HNCs).
Methods: We utilized whole exome sequencing to analyze somatic nucleotide (SNVs) and copy number variants (CNVs) among a unique set of 51 primary HPV-related OPSCCs, including 35 that did not recur and 16 that recurred. We evaluated 12 metachronous recurrent OPSCCs (7 with paired primary OPSCCs) and 33 primary HPV-unrelated oral cavity and OPSCCs.
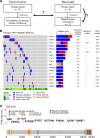
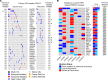
Results: KMT2D was the most frequently mutated gene among primary HPV-related OPSCCs (n = 51; 14%) and among metachronous recurrent OPSCCs (n = 12; 42%). Primary HPV-related OPSCCs that recurred shared a genomic landscape with primary HPV-related OPSCCs that did not recur. However, TSC2, BRIP1, NBN, and NFE2L2 mutations occurred in primary OPSCCs that recurred but not in those that did not recur. Moreover, primary HPV-related OPSCCs that recur harbor features of HPV-unrelated HNCs, notably including MAPK, JAK/STAT, and differentiation signaling pathway aberrations. Metachronous recurrent OPSCCs shared a genomic landscape with HPV-unrelated HNCs, including a high frequency of TP53, CASP8, FAT1, HLA-A, AJUBA, and NSD1 genomic alterations.
Conclusion: Overall, primary HPV-related OPSCCs that recur share a genomic landscape with nonrecurrent OPSCCs. Metachronous recurrent OPSCCs share genomic features with HPV-negative HNCs. These data aim to guide future deescalation endeavors and functional experiments.
Funding: This study is supported by the American Cancer Society (RSG TBG-123653), funding support for RAH (T32DC00018, Research Training in Otolaryngology, University of Washington), funds to EM from Seattle Translational Tumor Research (Fred Hutchinson Cancer Research Center), and center funds from the Fred Hutchinson Cancer Research Center to EM. UD is supported by the Department of Veterans Affairs, Biomedical Laboratory Research and Development (BLR&D), grant IO1-oo23456, and funds from the Pittsburgh Foundation and PNC Foundation.
Keywords: Cancer; Head & neck cancer; Molecular diagnosis; Oncology.
Conflict of interest statement
Figures



Similar articles
-
Mutation patterns in recurrent and/or metastatic oropharyngeal squamous cell carcinomas in relation to human papillomavirus status.Cancer Med. 2021 Feb;10(4):1347-1356. doi: 10.1002/cam4.3741. Epub 2021 Feb 1. Cancer Med. 2021. PMID: 33527763 Free PMC article.
-
Molecular subclassification determined by human papillomavirus and epidermal growth factor receptor status is associated with the prognosis of oropharyngeal squamous cell carcinoma.Hum Pathol. 2016 Apr;50:51-61. doi: 10.1016/j.humpath.2015.11.001. Epub 2015 Nov 17. Hum Pathol. 2016. PMID: 26997438
-
HPV+ oropharyngeal squamous cell carcinomas from patients with two tumors display synchrony of viral genomes yet discordant mutational profiles and signatures.Carcinogenesis. 2021 Feb 11;42(1):14-20. doi: 10.1093/carcin/bgaa111. Carcinogenesis. 2021. PMID: 33075810 Free PMC article.
-
Prognostic Significance of the Microenvironment in Human Papillomavirus Oropharyngeal Carcinoma: A Systematic Review.Laryngoscope. 2024 Apr;134(4):1507-1516. doi: 10.1002/lary.31010. Epub 2023 Aug 29. Laryngoscope. 2024. PMID: 37642393 Review.
-
The New Face of Head and Neck Cancer: The HPV Epidemic.Oncology (Williston Park). 2015 Sep;29(9):616-26. Oncology (Williston Park). 2015. PMID: 26384796 Review.
Cited by
-
Utility of plasma circulating tumor DNA and tumor DNA profiles in head and neck squamous cell carcinoma.Sci Rep. 2022 Jun 4;12(1):9316. doi: 10.1038/s41598-022-13417-5. Sci Rep. 2022. PMID: 35661138 Free PMC article.
-
Pyk2/FAK Signaling Is Upregulated in Recurrent Glioblastoma Tumors in a C57BL/6/GL261 Glioma Implantation Model.Int J Mol Sci. 2023 Aug 30;24(17):13467. doi: 10.3390/ijms241713467. Int J Mol Sci. 2023. PMID: 37686276 Free PMC article.
-
Potential of Pembrolizumab in Metastatic or Recurrent Head and Neck Cancer: Evidence to Date.Onco Targets Ther. 2020 Apr 9;13:3047-3059. doi: 10.2147/OTT.S196252. eCollection 2020. Onco Targets Ther. 2020. PMID: 32308436 Free PMC article. Review.
-
A benchmark for oncologic outcomes and model for lethal recurrence risk after transoral robotic resection of HPV-related oropharyngeal cancers.Oral Oncol. 2022 Apr;127:105798. doi: 10.1016/j.oraloncology.2022.105798. Epub 2022 Mar 1. Oral Oncol. 2022. PMID: 35245888 Free PMC article.
-
Mutation patterns in recurrent and/or metastatic oropharyngeal squamous cell carcinomas in relation to human papillomavirus status.Cancer Med. 2021 Feb;10(4):1347-1356. doi: 10.1002/cam4.3741. Epub 2021 Feb 1. Cancer Med. 2021. PMID: 33527763 Free PMC article.
References
-
- McQuillan G, Kruszon-Moran D, Markowitz LE, Unger ER, Paulose-Ram R. Prevalence of HPV in Adults Aged 18-69: United States, 2011-2014. NCHS Data Brief. 2017;(280):1–8. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

