Candida albicans-Induced NETosis Is Independent of Peptidylarginine Deiminase 4
- PMID: 30038623
- PMCID: PMC6046457
- DOI: 10.3389/fimmu.2018.01573
Candida albicans-Induced NETosis Is Independent of Peptidylarginine Deiminase 4
Abstract
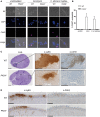
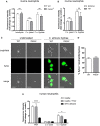
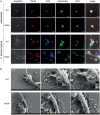
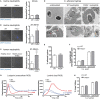
Neutrophils are the most abundant innate immune cells and the first line of defense against many pathogenic microbes, including the human fungal pathogen Candida albicans. Among the neutrophils' arsenal of effector functions, neutrophil extracellular traps (NETs) are thought to be of particular importance for trapping and killing the large fungal filaments by means of their web-like structures that consist of chromatin fibers decorated with proteolytic enzymes and host defense proteins. Peptidylarginine deiminase 4 (PAD4)-mediated citrullination of histones in activated neutrophils correlates with chromatin decondensation and extrusion and is widely accepted to act as an integral process of NET induction (NETosis). However, the requirement of PAD4-mediated histone citrullination for NET release during C. albicans infection remains unclear. In this study, we show that although PAD4-dependent neutrophil histone citrullination is readily induced by C. albicans, PAD4 is dispensable for NETosis in response to the fungus and other common NET-inducing stimuli. Moreover, PAD4 is not required for antifungal immunity during mucosal and systemic C. albicans infection. Our results demonstrate that PAD4 is dispensable for C. albicans-induced NETosis, and they highlight the limitations of using histone citrullination as a marker for NETs and PAD4-/- mice as a model of NET-deficiency.
Keywords: Candida albicans; host–fungus interaction; neutrophil extracellular traps; peptidylarginine deiminase 4.
Figures
Similar articles
-
NETosis proceeds by cytoskeleton and endomembrane disassembly and PAD4-mediated chromatin decondensation and nuclear envelope rupture.Proc Natl Acad Sci U S A. 2020 Mar 31;117(13):7326-7337. doi: 10.1073/pnas.1909546117. Epub 2020 Mar 13. Proc Natl Acad Sci U S A. 2020. PMID: 32170015 Free PMC article.
-
PAD4 mediated histone hypercitrullination induces heterochromatin decondensation and chromatin unfolding to form neutrophil extracellular trap-like structures.Front Immunol. 2012 Oct 4;3:307. doi: 10.3389/fimmu.2012.00307. eCollection 2012. Front Immunol. 2012. PMID: 23060885 Free PMC article.
-
Peptidylarginine deiminase 2 is required for tumor necrosis factor alpha-induced citrullination and arthritis, but not neutrophil extracellular trap formation.J Autoimmun. 2017 Jun;80:39-47. doi: 10.1016/j.jaut.2017.01.006. Epub 2017 Feb 7. J Autoimmun. 2017. PMID: 28188029 Free PMC article.
-
PAD4 takes charge during neutrophil activation: Impact of PAD4 mediated NET formation on immune-mediated disease.J Thromb Haemost. 2021 Jul;19(7):1607-1617. doi: 10.1111/jth.15313. Epub 2021 May 12. J Thromb Haemost. 2021. PMID: 33773016 Free PMC article. Review.
-
ROLE OF PEPTIDYLARGININE DEIMINASE AND NEUTROPHIL EXTRACELLULAR TRAPS IN INJURIES: FUTURE NOVEL DIAGNOSTICS AND THERAPEUTIC TARGETS.Shock. 2023 Feb 1;59(2):247-255. doi: 10.1097/SHK.0000000000002052. Epub 2023 Jan 5. Shock. 2023. PMID: 36597759 Free PMC article. Review.
Cited by
-
A Narrative Review: The Role of NETs in Acute Respiratory Distress Syndrome/Acute Lung Injury.Int J Mol Sci. 2024 Jan 25;25(3):1464. doi: 10.3390/ijms25031464. Int J Mol Sci. 2024. PMID: 38338744 Free PMC article. Review.
-
Role of Extracellular Trap Release During Bacterial and Viral Infection.Front Microbiol. 2022 Jan 26;13:798853. doi: 10.3389/fmicb.2022.798853. eCollection 2022. Front Microbiol. 2022. PMID: 35154050 Free PMC article. Review.
-
Candida albicans triggers NADPH oxidase-independent neutrophil extracellular traps through dectin-2.PLoS Pathog. 2019 Nov 6;15(11):e1008096. doi: 10.1371/journal.ppat.1008096. eCollection 2019 Nov. PLoS Pathog. 2019. PMID: 31693704 Free PMC article.
-
Reduced Anti-Histone Antibodies and Increased Risk of Rheumatoid Arthritis Associated with a Single Nucleotide Polymorphism in PADI4 in North Americans.Int J Mol Sci. 2019 Jun 25;20(12):3093. doi: 10.3390/ijms20123093. Int J Mol Sci. 2019. PMID: 31242568 Free PMC article.
-
A Synergy Between Endotoxin and (1→3)-Beta-D-Glucan Enhanced Neutrophil Extracellular Traps in Candida Administered Dextran Sulfate Solution Induced Colitis in FcGRIIB-/- Lupus Mice, an Impact of Intestinal Fungi in Lupus.J Inflamm Res. 2021 Jun 1;14:2333-2352. doi: 10.2147/JIR.S305225. eCollection 2021. J Inflamm Res. 2021. PMID: 34103965 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

