Evaluating Anti-CD32b F(ab) Conformation Using Molecular Dynamics and Small-Angle X-Ray Scattering
- PMID: 30021105
- PMCID: PMC6050753
- DOI: 10.1016/j.bpj.2018.03.040
Evaluating Anti-CD32b F(ab) Conformation Using Molecular Dynamics and Small-Angle X-Ray Scattering
Abstract
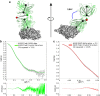
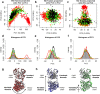
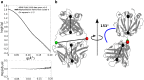
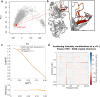
Complementary strategies of small-angle x-ray scattering (SAXS) and crystallographic analysis are often used to determine atomistic three-dimensional models of macromolecules and their variability in solution. This combination of techniques is particularly valuable when applied to macromolecular complexes to detect changes within the individual binding partners. Here, we determine the x-ray crystallographic structure of a F(ab) fragment in complex with CD32b, the only inhibitory Fc-γ receptor in humans, and compare the structure of the F(ab) from the crystal complex to SAXS data for the F(ab) alone in solution. We investigate changes in F(ab) structure by predicting theoretical scattering profiles for atomistic structures extracted from molecular dynamics (MD) simulations of the F(ab) and assessing the agreement of these structures to our experimental SAXS data. Through principal component analysis, we are able to extract principal motions observed during the MD trajectory and evaluate the influence of these motions on the agreement of structures to the F(ab) SAXS data. Changes in the F(ab) elbow angle were found to be important to reach agreement with the experimental data; however, further discrepancies were apparent between our F(ab) structure from the crystal complex and SAXS data. By analyzing multiple MD structures observed in similar regions of the principal component analysis, we were able to pinpoint these discrepancies to a specific loop region in the F(ab) heavy chain. This method, therefore, not only allows determination of global changes but also allows identification of localized motions important for determining the agreement between atomistic structures and SAXS data. In this particular case, the findings allowed us to discount the hypothesis that structural changes were induced upon complex formation, a significant find informing the drug development process. The methodology described here is generally applicable to deconvolute global and local changes of macromolecular structures and is well suited to other systems.
Copyright © 2018 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Hybrid Methods for Modeling Protein Structures Using Molecular Dynamics Simulations and Small-Angle X-Ray Scattering Data.Adv Exp Med Biol. 2018;1105:237-258. doi: 10.1007/978-981-13-2200-6_15. Adv Exp Med Biol. 2018. PMID: 30617833 Review.
-
Crystal structures and molecular dynamics simulations of a humanised antibody fragment at acidic to basic pH.Sci Rep. 2023 Sep 28;13(1):16281. doi: 10.1038/s41598-023-42698-7. Sci Rep. 2023. PMID: 37770469 Free PMC article.
-
An Expanded Conformation of an Antibody Fab Region by X-Ray Scattering, Molecular Dynamics, and smFRET Identifies an Aggregation Mechanism.J Mol Biol. 2019 Mar 29;431(7):1409-1425. doi: 10.1016/j.jmb.2019.02.009. Epub 2019 Feb 16. J Mol Biol. 2019. PMID: 30776431
-
Molecular Dynamics Simulations Combined with Nuclear Magnetic Resonance and/or Small-Angle X-ray Scattering Data for Characterizing Intrinsically Disordered Protein Conformational Ensembles.J Chem Inf Model. 2019 May 28;59(5):1743-1758. doi: 10.1021/acs.jcim.8b00928. Epub 2019 Mar 18. J Chem Inf Model. 2019. PMID: 30840442 Review.
-
Characterization of the NISTmAb Reference Material using small-angle scattering and molecular simulation : Part I: Dilute protein solutions.Anal Bioanal Chem. 2018 Mar;410(8):2141-2159. doi: 10.1007/s00216-018-0868-2. Epub 2018 Feb 9. Anal Bioanal Chem. 2018. PMID: 29423600
Cited by
-
The Structural and Functional Diversity of Intrinsically Disordered Regions in Transmembrane Proteins.J Membr Biol. 2019 Oct;252(4-5):273-292. doi: 10.1007/s00232-019-00069-2. Epub 2019 May 28. J Membr Biol. 2019. PMID: 31139867 Review.
-
Assessing and Engineering Antibody Stability Using Experimental and Computational Methods.Methods Mol Biol. 2023;2552:165-197. doi: 10.1007/978-1-0716-2609-2_9. Methods Mol Biol. 2023. PMID: 36346592
-
Rigid crosslinking of the CD3 complex leads to superior T cell stimulation.Front Immunol. 2024 Aug 30;15:1434463. doi: 10.3389/fimmu.2024.1434463. eCollection 2024. Front Immunol. 2024. PMID: 39281668 Free PMC article.
-
Molecular Dynamics Simulations Reveal Interactions of an IgG1 Antibody With Selected Fc Receptors.Front Chem. 2021 Jul 2;9:705931. doi: 10.3389/fchem.2021.705931. eCollection 2021. Front Chem. 2021. PMID: 34277572 Free PMC article.
-
Molecular Dynamics Simulations Reveal How Competing Protein-Surface Interactions for Glycine, Citrate, and Water Modulate Stability in Antibody Fragment Formulations.Mol Pharm. 2024 Nov 4;21(11):5497-5509. doi: 10.1021/acs.molpharmaceut.4c00332. Epub 2024 Oct 21. Mol Pharm. 2024. PMID: 39431440 Free PMC article.
References
-
- Scott A.M., Wolchok J.D., Old L.J. Antibody therapy of cancer. Nat. Rev. Cancer. 2012;12:278–287. - PubMed
-
- Chan A.C., Carter P.J. Therapeutic antibodies for autoimmunity and inflammation. Nat. Rev. Immunol. 2010;10:301–316. - PubMed
-
- Nimmerjahn F., Ravetch J.V. Fcgamma receptors as regulators of immune responses. Nat. Rev. Immunol. 2008;8:34–47. - PubMed
-
- Yu X., Marshall M.J.E., Crispin M. Improving antibody-based cancer therapeutics through glycan engineering. BioDrugs. 2017;31:151–166. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

