A simple supported tubulated bilayer system for evaluating protein-mediated membrane remodeling
- PMID: 30012406
- PMCID: PMC6103888
- DOI: 10.1016/j.chemphyslip.2018.06.002
A simple supported tubulated bilayer system for evaluating protein-mediated membrane remodeling
Abstract
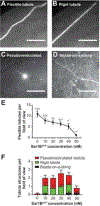
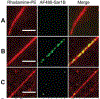
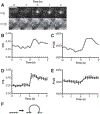
Fusion and fission of cellular membranes involve dramatic, protein-mediated changes in membrane curvature. Many of the experimental methods useful for investigating curvature sensing or generation require specialized equipment. We have developed a system based on supported lipid bilayers (SLBs) in which lipid tubules are simple to produce and several types of membrane remodeling events can be readily imaged using widely available instrumentation (e.g., tubule fission and/or membrane budding). Briefly, high ionic strength during lipid bilayer deposition results in incorporation of excess lipids in the SLB. After sequentially washing with water and physiological ionic strength buffer solutions, lipid tubules form spontaneously. We find that tubule formation results from solution-dependent spreading of the SLB; washing from water into physiological ionic strength buffer solution leads to expansion of the bilayer and formation of tubules. Conversely, washing from physiological buffer into water results in contraction of the membrane and loss of tubules. We demonstrate the utility of these supported tubulated bilayers, termed "STuBs," with an investigation of Sar1B, a small Ras family G-protein known to influence membrane curvature. The addition of Sar1B to STuBs results in dramatic changes in tubule topology and eventual tubule fission. Overall, STuBs are a simple experimental system, useful for monitoring protein-mediated effects on membrane topology in real time, under physiologically relevant conditions.
Keywords: Endocytosis; Exocytosis; Membrane fission; Supported bilayer; Vesicle budding.
Copyright © 2018 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors have no competing interests to declare, other than the grant support noted above.
Figures



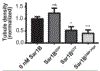
Similar articles
-
Supported bilayers with excess membrane reservoir: a template for reconstituting membrane budding and fission.Biophys J. 2010 Jul 21;99(2):517-25. doi: 10.1016/j.bpj.2010.04.036. Biophys J. 2010. PMID: 20643070 Free PMC article.
-
Sensing Ability and Formation Criterion of Fluid Supported Lipid Bilayer Coated Graphene Field-Effect Transistors.ACS Sens. 2019 Apr 26;4(4):892-899. doi: 10.1021/acssensors.8b01623. Epub 2019 Mar 13. ACS Sens. 2019. PMID: 30817891
-
The importance of membrane defects-lessons from simulations.Acc Chem Res. 2014 Aug 19;47(8):2244-51. doi: 10.1021/ar4002729. Epub 2014 Jun 3. Acc Chem Res. 2014. PMID: 24892900
-
Dynamin: functional design of a membrane fission catalyst.Annu Rev Cell Dev Biol. 2011;27:79-105. doi: 10.1146/annurev-cellbio-100109-104016. Epub 2011 May 18. Annu Rev Cell Dev Biol. 2011. PMID: 21599493 Review.
-
Protein-lipid interplay in fusion and fission of biological membranes.Annu Rev Biochem. 2003;72:175-207. doi: 10.1146/annurev.biochem.72.121801.161504. Annu Rev Biochem. 2003. PMID: 14527322 Review.
Cited by
-
Passive and reversible area regulation of supported lipid bilayers in response to fluid flow.Biophys J. 2023 Jun 6;122(11):2242-2255. doi: 10.1016/j.bpj.2023.01.012. Epub 2023 Jan 13. Biophys J. 2023. PMID: 36639867 Free PMC article.
-
Phosphatidic Acid Accumulates at Areas of Curvature in Tubulated Lipid Bilayers and Liposomes.Biomolecules. 2022 Nov 17;12(11):1707. doi: 10.3390/biom12111707. Biomolecules. 2022. PMID: 36421720 Free PMC article.
-
COPII-mediated trafficking at the ER/ERGIC interface.Traffic. 2019 Jul;20(7):491-503. doi: 10.1111/tra.12654. Epub 2019 May 30. Traffic. 2019. PMID: 31059169 Free PMC article. Review.
-
Coupling liquid phases in 3D condensates and 2D membranes: Successes, challenges, and tools.Biophys J. 2024 Jun 4;123(11):1329-1341. doi: 10.1016/j.bpj.2023.12.023. Epub 2023 Dec 29. Biophys J. 2024. PMID: 38160256 Review.
-
Single-molecule phospholipase A2 becomes processive on melittin-induced membrane deformations.Biophys J. 2022 Apr 19;121(8):1417-1423. doi: 10.1016/j.bpj.2022.03.019. Epub 2022 Mar 18. Biophys J. 2022. PMID: 35314142 Free PMC article.
References
-
- ADOLF F, HERRMANN A, HELLWIG A, BECK R, BRUGGER B & WIELAND FT 2013. Scission of COPI and COPII vesicles is independent of GTP hydrolysis. Traffic, 14, 922–32. - PubMed
-
- ALNAAS AA, MOON CL, ALTON M, REED SM & KNOWLES MK 2017. Conformational Changes in C-Reactive Protein Affect Binding to Curved Membranes in a Lipid Bilayer Model of the Apoptotic Cell Surface. The Journal of Physical Chemistry B, 121, 2631–2639. - PubMed
-
- ANDERSON TH, MIN Y, WEIRICH KL, ZENG H, FYGENSON D & ISRAELACHVILI JN 2009. Formation of supported bilayers on silica substrates. Langmuir, 25, 6997–7005. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

