Review and Meta-Analyses of TAAR1 Expression in the Immune System and Cancers
- PMID: 29997511
- PMCID: PMC6029583
- DOI: 10.3389/fphar.2018.00683
Review and Meta-Analyses of TAAR1 Expression in the Immune System and Cancers
Abstract
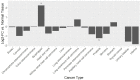
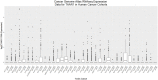
Since its discovery in 2001, the major focus of TAAR1 research has been on its role in monoaminergic regulation, drug-induced reward and psychiatric conditions. More recently, TAAR1 expression and functionality in immune system regulation and immune cell activation has become a topic of emerging interest. Here, we review the immunologically-relevant TAAR1 literature and incorporate open-source expression and cancer survival data meta-analyses. We provide strong evidence for TAAR1 expression in the immune system and cancers revealed through NCBI GEO datamining and discuss its regulation in a spectrum of immune cell types as well as in numerous cancers. We discuss connections and logical directions for further study of TAAR1 in immunological function, and its potential role as a mediator or modulator of immune dysregulation, immunological effects of psychostimulant drugs of abuse, and cancer progression.
Keywords: B-cell; PBMC; T-cell; astrocyte; granulocyte; lymphocyte; microglia; platelet.
Figures

Similar articles
-
Methamphetamine induces trace amine-associated receptor 1 (TAAR1) expression in human T lymphocytes: role in immunomodulation.J Leukoc Biol. 2016 Jan;99(1):213-23. doi: 10.1189/jlb.4A0814-395RR. Epub 2015 Aug 24. J Leukoc Biol. 2016. PMID: 26302754 Free PMC article.
-
Trace Amine Associated Receptor 1 (TAAR1) Modulation of Food Reward.Front Pharmacol. 2018 Feb 27;9:129. doi: 10.3389/fphar.2018.00129. eCollection 2018. Front Pharmacol. 2018. PMID: 29535626 Free PMC article. Review.
-
Trace amine associated receptor 1 (TAAR1) modulators: a patent review (2010-present).Expert Opin Ther Pat. 2020 Feb;30(2):137-145. doi: 10.1080/13543776.2020.1708900. Epub 2019 Dec 25. Expert Opin Ther Pat. 2020. PMID: 31865810 Review.
-
TAAR1 in Addiction: Looking Beyond the Tip of the Iceberg.Front Pharmacol. 2018 Mar 27;9:279. doi: 10.3389/fphar.2018.00279. eCollection 2018. Front Pharmacol. 2018. PMID: 29636691 Free PMC article. Review.
-
The Case for TAAR1 as a Modulator of Central Nervous System Function.Front Pharmacol. 2018 Jan 10;8:987. doi: 10.3389/fphar.2017.00987. eCollection 2017. Front Pharmacol. 2018. PMID: 29375386 Free PMC article. Review.
Cited by
-
Critical Aspects Affecting Cannabidiol Oral Bioavailability and Metabolic Elimination, and Related Clinical Implications.CNS Drugs. 2020 Aug;34(8):795-800. doi: 10.1007/s40263-020-00741-5. CNS Drugs. 2020. PMID: 32504461 Review.
-
Modelling of p-tyramine transport across human intestinal epithelial cells predicts the presence of additional transporters.Front Physiol. 2022 Nov 10;13:1009320. doi: 10.3389/fphys.2022.1009320. eCollection 2022. Front Physiol. 2022. PMID: 36505075 Free PMC article.
-
Unveiling the enigmatic roles of basophils in HIV infection: A narrative review.Medicine (Baltimore). 2024 Nov 1;103(44):e40384. doi: 10.1097/MD.0000000000040384. Medicine (Baltimore). 2024. PMID: 39496030 Free PMC article. Review.
-
An Integrative Metabolomic and Network Pharmacology Study Revealing the Regulating Properties of Xihuang Pill That Improves Anlotinib Effects in Lung Cancer.Front Oncol. 2021 Aug 9;11:697247. doi: 10.3389/fonc.2021.697247. eCollection 2021. Front Oncol. 2021. PMID: 34434895 Free PMC article.
-
TAAR1 Regulates Purinergic-induced TNF Secretion from Peripheral, But Not CNS-resident, Macrophages.J Neuroimmune Pharmacol. 2023 Jun;18(1-2):100-111. doi: 10.1007/s11481-022-10053-8. Epub 2022 Nov 16. J Neuroimmune Pharmacol. 2023. PMID: 36380156
References
-
- An D., Kim K., Lu W. (2014). Defective Entry into Mitosis 1 (Dim1) negatively regulates osteoclastogenesis by inhibiting the expression of Nuclear Factor of Activated T-cells, Cytoplasmic, Calcineurin-dependent 1 (NFATc1). J. Biol. Chem. 289, 24366–24373. 10.1074/jbc.m114.563817 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

