Histone Citrullination Represses MicroRNA Expression, Resulting in Increased Oncogene mRNAs in Somatolactotrope Cells
- PMID: 29987187
- PMCID: PMC6146832
- DOI: 10.1128/MCB.00084-18
Histone Citrullination Represses MicroRNA Expression, Resulting in Increased Oncogene mRNAs in Somatolactotrope Cells
Abstract
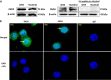
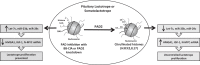
Peptidylarginine deiminase (PAD) enzymes convert histone arginine residues into citrulline to modulate chromatin organization and gene expression. Although PADs are expressed in anterior pituitary gland cells, their functional role and expression in pituitary adenomas are unknown. To begin to address these issues, we first examined normal human pituitaries and pituitary adenomas and found that PAD2, PAD4, and citrullinated histones are highest in prolactinomas and somatoprolactinomas. In the somatoprolactinoma-derived GH3 cell line, PADs citrullinate histone H3, which is attenuated by a pan-PAD inhibitor. RNA sequencing and chromatin immunoprecipitation (ChIP) studies show that the expression of microRNAs (miRNAs) let-7c-2, 23b, and 29c is suppressed by histone citrullination. Our studies demonstrate that these miRNAs directly target the mRNA of the oncogenes encoding HMGA, insulin-like growth factor 1 (IGF-1), and N-MYC, which are highly implicated in human prolactinoma/somatoprolactinoma pathogenesis. Our results are the first to define a direct role for PAD-catalyzed histone citrullination in miRNA expression, which may underlie the etiology of prolactinoma and somatoprolactinoma tumors through regulation of oncogene expression.
Keywords: citrullination; epigenetics; miRNA; oncogenes; prolactinoma.
Copyright © 2018 American Society for Microbiology.
Figures



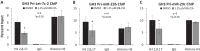

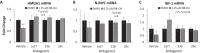

Similar articles
-
Citrullination regulates the expression of insulin-like growth factor-binding protein 1 (IGFBP1) in ovine uterine luminal epithelial cells.Reproduction. 2017 Jan;153(1):1-10. doi: 10.1530/REP-16-0494. Epub 2016 Oct 10. Reproduction. 2017. PMID: 29565015 Free PMC article.
-
GnRH Stimulates Peptidylarginine Deiminase Catalyzed Histone Citrullination in Gonadotrope Cells.Mol Endocrinol. 2016 Oct;30(10):1081-1091. doi: 10.1210/me.2016-1085. Epub 2016 Sep 7. Mol Endocrinol. 2016. PMID: 27603413 Free PMC article.
-
Peptidylarginine deiminase enzymes and citrullinated proteins in female reproductive physiology and associated diseases†.Biol Reprod. 2022 Dec 10;107(6):1395-1410. doi: 10.1093/biolre/ioac173. Biol Reprod. 2022. PMID: 36087287 Free PMC article. Review.
-
Protein Citrullination by Peptidyl Arginine Deiminase/Arginine Deiminase Homologs in Members of the Human Microbiota and Its Recognition by Anti-Citrullinated Protein Antibodies.Int J Mol Sci. 2024 May 10;25(10):5192. doi: 10.3390/ijms25105192. Int J Mol Sci. 2024. PMID: 38791230 Free PMC article.
-
The roles of PAD2- and PAD4-mediated protein citrullination catalysis in cancers.Int J Cancer. 2021 Jan 15;148(2):267-276. doi: 10.1002/ijc.33205. Epub 2020 Aug 8. Int J Cancer. 2021. PMID: 33459350 Review.
Cited by
-
Multiomics Approach to Acromegaly: Unveiling Translational Insights for Precision Medicine.Endocrinol Metab (Seoul). 2023 Oct;38(5):463-471. doi: 10.3803/EnM.2023.1820. Epub 2023 Oct 13. Endocrinol Metab (Seoul). 2023. PMID: 37828709 Free PMC article. Review.
-
Unresolved Complexity in the Gene Regulatory Network Underlying EMT.Front Oncol. 2020 May 12;10:554. doi: 10.3389/fonc.2020.00554. eCollection 2020. Front Oncol. 2020. PMID: 32477926 Free PMC article. Review.
-
Inhibiting PAD2 enhances the anti-tumor effect of docetaxel in tamoxifen-resistant breast cancer cells.J Exp Clin Cancer Res. 2019 Oct 10;38(1):414. doi: 10.1186/s13046-019-1404-8. J Exp Clin Cancer Res. 2019. PMID: 31601253 Free PMC article.
-
The virtues and vices of protein citrullination.R Soc Open Sci. 2022 Jun 8;9(6):220125. doi: 10.1098/rsos.220125. eCollection 2022 Jun. R Soc Open Sci. 2022. PMID: 35706669 Free PMC article. Review.
-
Progression on Citrullination of Proteins in Gastrointestinal Cancers.Front Oncol. 2019 Jan 23;9:15. doi: 10.3389/fonc.2019.00015. eCollection 2019. Front Oncol. 2019. PMID: 30740359 Free PMC article. Review.
References
-
- Wang Y, Wysocka J, Sayegh J, Lee Y-H, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, Roeder RG, Clarke S, Stallcup MR, Allis CD, Coonrod SA. 2004. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 306:279–283. doi:10.1126/science.1101400. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
