Identification of a novel tRNA wobble uridine modifying activity in the biosynthesis of 5-methoxyuridine
- PMID: 29982645
- PMCID: PMC6158493
- DOI: 10.1093/nar/gky592
Identification of a novel tRNA wobble uridine modifying activity in the biosynthesis of 5-methoxyuridine
Abstract
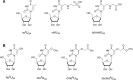
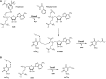
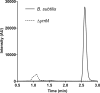
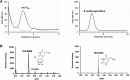
Derivatives of 5-hydroxyuridine (ho5U), such as 5-methoxyuridine (mo5U) and 5-oxyacetyluridine (cmo5U), are ubiquitous modifications of the wobble position of bacterial tRNA that are believed to enhance translational fidelity by the ribosome. In gram-negative bacteria, the last step in the biosynthesis of cmo5U from ho5U involves the unique metabolite carboxy S-adenosylmethionine (Cx-SAM) and the carboxymethyl transferase CmoB. However, the equivalent position in the tRNA of Gram-positive bacteria is instead mo5U, where the methyl group is derived from SAM and installed by an unknown methyltransferase. By utilizing a cmoB-deficient strain of Escherichia coli as a host and assaying for the formation of mo5U in total RNA isolates with methyltransferases of unknown function from Bacillus subtilis, we found that this modification is installed by the enzyme TrmR (formerly known as YrrM). Furthermore, X-ray crystal structures of TrmR with and without the anticodon stemloop of tRNAAla have been determined, which provide insight into both sequence and structure specificity in the interactions of TrmR with tRNA.
Figures
Similar articles
-
On the biosynthesis of 5-methoxyuridine and uridine-5-oxyacetic acid in specific procaryotic transfer RNAs.Nucleic Acids Res. 1978 Apr;5(4):1273-81. doi: 10.1093/nar/5.4.1273. Nucleic Acids Res. 1978. PMID: 418384 Free PMC article.
-
Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function.Nature. 2013 Jun 6;498(7452):123-6. doi: 10.1038/nature12180. Epub 2013 May 15. Nature. 2013. PMID: 23676670 Free PMC article.
-
Structural snapshots of CmoB in various states during wobble uridine modification of tRNA.Biochem Biophys Res Commun. 2021 Jan 1;534:604-609. doi: 10.1016/j.bbrc.2020.11.033. Epub 2020 Nov 17. Biochem Biophys Res Commun. 2021. PMID: 33213836
-
Stereochemical mechanisms of tRNA methyltransferases.FEBS Lett. 2010 Jan 21;584(2):278-86. doi: 10.1016/j.febslet.2009.11.075. FEBS Lett. 2010. PMID: 19944101 Free PMC article. Review.
-
Tied up in knots: Untangling substrate recognition by the SPOUT methyltransferases.J Biol Chem. 2022 Oct;298(10):102393. doi: 10.1016/j.jbc.2022.102393. Epub 2022 Aug 18. J Biol Chem. 2022. PMID: 35988649 Free PMC article. Review.
Cited by
-
Unique anticodon loop conformation with the flipped-out wobble nucleotide in the crystal structure of unbound tRNAVal.RNA. 2021 Nov;27(11):1330-1338. doi: 10.1261/rna.078863.121. Epub 2021 Jul 27. RNA. 2021. PMID: 34315814 Free PMC article.
-
Survey and Validation of tRNA Modifications and Their Corresponding Genes in Bacillus subtilis sp Subtilis Strain 168.Biomolecules. 2020 Jun 30;10(7):977. doi: 10.3390/biom10070977. Biomolecules. 2020. PMID: 32629984 Free PMC article.
-
Functions of Bacterial tRNA Modifications: From Ubiquity to Diversity.Trends Microbiol. 2021 Jan;29(1):41-53. doi: 10.1016/j.tim.2020.06.010. Epub 2020 Jul 25. Trends Microbiol. 2021. PMID: 32718697 Free PMC article. Review.
-
Identification and comparison of protein composition of biofilms in response to EGCG from Enterococcus faecalis and Staphylococcus lugdunensis, which showed opposite patterns in biofilm-forming abilities.Biofilm. 2024 Oct 24;8:100232. doi: 10.1016/j.bioflm.2024.100232. eCollection 2024 Dec. Biofilm. 2024. PMID: 39555139 Free PMC article.
-
Identification and Characterization of Genes Required for 5-Hydroxyuridine Synthesis in Bacillus subtilis and Escherichia coli tRNA.J Bacteriol. 2019 Sep 20;201(20):e00433-19. doi: 10.1128/JB.00433-19. Print 2019 Oct 15. J Bacteriol. 2019. PMID: 31358606 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

