A Muscle-Specific Enhancer RNA Mediates Cohesin Recruitment and Regulates Transcription In trans
- PMID: 29979962
- PMCID: PMC6082425
- DOI: 10.1016/j.molcel.2018.06.008
A Muscle-Specific Enhancer RNA Mediates Cohesin Recruitment and Regulates Transcription In trans
Abstract
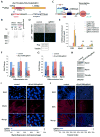
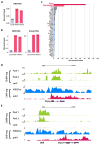
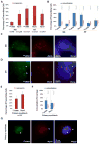
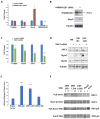
The enhancer regions of the myogenic master regulator MyoD give rise to at least two enhancer RNAs. Core enhancer eRNA (CEeRNA) regulates transcription of the adjacent MyoD gene, whereas DRReRNA affects expression of Myogenin in trans. We found that DRReRNA is recruited at the Myogenin locus, where it colocalizes with Myogenin nascent transcripts. DRReRNA associates with the cohesin complex, and this association correlates with its transactivating properties. Despite being expressed in undifferentiated cells, cohesin is not loaded on Myogenin until the cells start expressing DRReRNA, which is then required for cohesin chromatin recruitment and maintenance. Functionally, depletion of either cohesin or DRReRNA reduces chromatin accessibility, prevents Myogenin activation, and hinders muscle cell differentiation. Thus, DRReRNA ensures spatially appropriate cohesin loading in trans to regulate gene expression.
Keywords: Myogenin; cohesin; enhancer RNA; muscle gene transcription; non-coding RNA.
Published by Elsevier Inc.
Conflict of interest statement
The authors declare no competing interests.
Figures

Comment in
-
An Enhancer-Derived RNA Muscles In to Regulate Myogenin In trans.Mol Cell. 2018 Jul 5;71(1):3-5. doi: 10.1016/j.molcel.2018.06.024. Mol Cell. 2018. PMID: 29979967
Similar articles
-
MUNC, an Enhancer RNA Upstream from the MYOD Gene, Induces a Subgroup of Myogenic Transcripts in trans Independently of MyoD.Mol Cell Biol. 2018 Sep 28;38(20):e00655-17. doi: 10.1128/MCB.00655-17. Print 2018 Oct 15. Mol Cell Biol. 2018. PMID: 30037979 Free PMC article.
-
An Enhancer-Derived RNA Muscles In to Regulate Myogenin In trans.Mol Cell. 2018 Jul 5;71(1):3-5. doi: 10.1016/j.molcel.2018.06.024. Mol Cell. 2018. PMID: 29979967
-
MUNC, a long noncoding RNA that facilitates the function of MyoD in skeletal myogenesis.Mol Cell Biol. 2015 Feb;35(3):498-513. doi: 10.1128/MCB.01079-14. Epub 2014 Nov 17. Mol Cell Biol. 2015. PMID: 25403490 Free PMC article.
-
eRNA binding produces tailored CBP activity profiles to regulate gene expression.RNA Biol. 2017 Dec 2;14(12):1655-1659. doi: 10.1080/15476286.2017.1353862. Epub 2017 Sep 26. RNA Biol. 2017. PMID: 28891741 Free PMC article. Review.
-
Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis.Semin Cell Dev Biol. 2017 Dec;72:19-32. doi: 10.1016/j.semcdb.2017.11.011. Epub 2017 Nov 15. Semin Cell Dev Biol. 2017. PMID: 29127046 Review.
Cited by
-
Current Status of Epitranscriptomic Marks Affecting lncRNA Structures and Functions.Noncoding RNA. 2022 Mar 28;8(2):23. doi: 10.3390/ncrna8020023. Noncoding RNA. 2022. PMID: 35447886 Free PMC article. Review.
-
Enhancer RNAs are an important regulatory layer of the epigenome.Nat Struct Mol Biol. 2020 Jun;27(6):521-528. doi: 10.1038/s41594-020-0446-0. Epub 2020 Jun 8. Nat Struct Mol Biol. 2020. PMID: 32514177 Free PMC article. Review.
-
Roles of super enhancers and enhancer RNAs in skeletal muscle development and disease.Cell Cycle. 2023 Mar;22(5):495-505. doi: 10.1080/15384101.2022.2129240. Epub 2022 Oct 2. Cell Cycle. 2023. PMID: 36184878 Free PMC article. Review.
-
Distinct MUNC lncRNA structural domains regulate transcription of different promyogenic factors.Cell Rep. 2022 Feb 15;38(7):110361. doi: 10.1016/j.celrep.2022.110361. Cell Rep. 2022. PMID: 35172143 Free PMC article.
-
The role of enhancer RNAs in epigenetic regulation of gene expression.Transcription. 2020 Feb;11(1):19-25. doi: 10.1080/21541264.2019.1698934. Epub 2019 Dec 11. Transcription. 2020. PMID: 31823686 Free PMC article. Review.
References
-
- Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H, Shiroishi T. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Developmental cell. 2009;16:47–57. - PubMed
-
- Asakura A, Lyons GE, Tapscott SJ. The regulation of MyoD gene expression: conserved elements mediate expression in embryonic axial muscle. Dev Biol. 1995;171:386–398. - PubMed
-
- Austenaa LM, Barozzi I, Simonatto M, Masella S, Della Chiara G, Ghisletti S, Curina A, de Wit E, Bouwman BA, de Pretis S, et al. Transcription of Mammalian cis-Regulatory Elements Is Restrained by Actively Enforced Early Termination. Molecular cell. 2015;60:460–474. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

