Comparative Chloroplast Genome Analyses of Species in Gentiana section Cruciata (Gentianaceae) and the Development of Authentication Markers
- PMID: 29976857
- PMCID: PMC6073106
- DOI: 10.3390/ijms19071962
Comparative Chloroplast Genome Analyses of Species in Gentiana section Cruciata (Gentianaceae) and the Development of Authentication Markers
Abstract
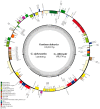
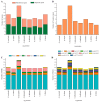
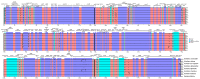
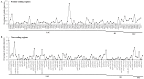
Gentiana section Cruciata is widely distributed across Eurasia at high altitudes, and some species in this section are used as traditional Chinese medicine. Accurate identification of these species is important for their utilization and conservation. Due to similar morphological and chemical characteristics, correct discrimination of these species still remains problematic. Here, we sequenced three complete chloroplast (cp) genomes (G. dahurica, G. siphonantha and G. officinalis). We further compared them with the previously published plastomes from sect. Cruciata and developed highly polymorphic molecular markers for species authentication. The eight cp genomes shared the highly conserved structure and contained 112 unique genes arranged in the same order, including 78 protein-coding genes, 30 tRNAs, and 4 rRNAs. We analyzed the repeats and nucleotide substitutions in these plastomes and detected several highly variable regions. We found that four genes (accD, clpP, matK and ycf1) were subject to positive selection, and sixteen InDel-variable loci with high discriminatory powers were selected as candidate barcodes. Our phylogenetic analyses based on plastomes further confirmed the monophyly of sect. Cruciata and primarily elucidated the phylogeny of Gentianales. This study indicated that cp genomes can provide more integrated information for better elucidating the phylogenetic pattern and improving discriminatory power during species authentication.
Keywords: Gentiana section Cruciata; chloroplast genome; molecular markers; species authentication.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


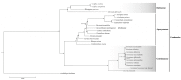
 in the phylogenetic tree indicated that the support value of branch is 100/100/1.0.
in the phylogenetic tree indicated that the support value of branch is 100/100/1.0.Similar articles
-
Chloroplast genome structures in Gentiana (Gentianaceae), based on three medicinal alpine plants used in Tibetan herbal medicine.Curr Genet. 2017 May;63(2):241-252. doi: 10.1007/s00294-016-0631-1. Epub 2016 Jul 15. Curr Genet. 2017. PMID: 27422574
-
[A strategy for identifying six species of Sect.Cruciata(Gentiana) in Gansu using DNA barcode sequences].Yao Xue Xue Bao. 2016 May;51(5):821-7. Yao Xue Xue Bao. 2016. PMID: 29878733 Chinese.
-
The complete chloroplast genome of Gentiana straminea (Gentianaceae), an endemic species to the Sino-Himalayan subregion.Gene. 2016 Feb 15;577(2):281-8. doi: 10.1016/j.gene.2015.12.005. Epub 2015 Dec 9. Gene. 2016. PMID: 26680100
-
Comparative Genomic and Phylogenetic Analysis of Forty Gentiana Chloroplast Genomes.Front Biosci (Landmark Ed). 2022 Aug 9;27(8):236. doi: 10.31083/j.fbl2708236. Front Biosci (Landmark Ed). 2022. PMID: 36042168
-
Comparative transcriptome analyses of three Gentiana species provides signals for the molecular footprints of selection effects and the phylogenetic relationships.Mol Genet Genomics. 2023 Mar;298(2):399-411. doi: 10.1007/s00438-022-01991-2. Epub 2023 Jan 2. Mol Genet Genomics. 2023. PMID: 36592219
Cited by
-
Comparative Analysis of Six Chloroplast Genomes in Chenopodium and Its Related Genera (Amaranthaceae): New Insights into Phylogenetic Relationships and the Development of Species-Specific Molecular Markers.Genes (Basel). 2023 Dec 6;14(12):2183. doi: 10.3390/genes14122183. Genes (Basel). 2023. PMID: 38137004 Free PMC article.
-
Lineage-specific plastid degradation in subtribe Gentianinae (Gentianaceae).Ecol Evol. 2021 Feb 22;11(7):3286-3299. doi: 10.1002/ece3.7281. eCollection 2021 Apr. Ecol Evol. 2021. PMID: 33841784 Free PMC article.
-
Complete chloroplast genome structure of four Ulmus species and Hemiptelea davidii and comparative analysis within Ulmaceae species.Sci Rep. 2022 Sep 24;12(1):15953. doi: 10.1038/s41598-022-20184-w. Sci Rep. 2022. PMID: 36153397 Free PMC article.
-
Comparative analysis of whole chloroplast genomes of three common species of Nekemias from vine tea.Sci Rep. 2024 Aug 17;14(1):19107. doi: 10.1038/s41598-024-69932-0. Sci Rep. 2024. PMID: 39154140 Free PMC article.
-
Evolutionary analysis of six chloroplast genomes from three Persea americana ecological races: Insights into sequence divergences and phylogenetic relationships.PLoS One. 2019 Sep 18;14(9):e0221827. doi: 10.1371/journal.pone.0221827. eCollection 2019. PLoS One. 2019. PMID: 31532782 Free PMC article.
References
-
- Zhang X.L., Wang Y.J., Ge X.J., Yuan Y.M., Yang H.L., Liu J.Q. Molecular phylogeny and biogeography of Gentiana sect. Cruciata (Gentianaceae) based on four chloroplast DNA datasets. Taxon. 2009;58:862–870.
-
- Ho T.N., Liu S.W. A Worldwide Monograph of Gentiana. Science Press; Beijing, China: 2001.
-
- Ho T.N., Pringle S.J. “Gentianaceae,” Flora of China. Volume 16. Science Press; Beijing, China: 1995. pp. 1–140.
-
- State Pharmacopoeia Commission of the PRC . Pharmacopoeia of P.R. China, Part 1. Chemical Industry Publishing House; Beijing, China: 2015. pp. 270–271.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

