The methyltransferase PRMT6 attenuates antiviral innate immunity by blocking TBK1-IRF3 signaling
- PMID: 29973649
- PMCID: PMC6804946
- DOI: 10.1038/s41423-018-0057-4
The methyltransferase PRMT6 attenuates antiviral innate immunity by blocking TBK1-IRF3 signaling
Erratum in
-
Correction to: The methyltransferase PRMT6 attenuates antiviral innate immunity by blocking TBK1-IRF3 signaling.Cell Mol Immunol. 2020 Feb;17(2):192. doi: 10.1038/s41423-019-0337-7. Cell Mol Immunol. 2020. PMID: 31776454 Free PMC article.
Abstract
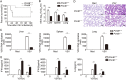
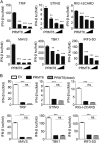
Protein arginine methyltransferases (PRMTs) play diverse biological roles and are specifically involved in immune cell development and inflammation. However, their role in antiviral innate immunity has not been elucidated. Viral infection triggers the TBK1-IRF3 signaling pathway to stimulate the production of type-I interferon, which mediates antiviral immunity. We performed a functional screen of the nine mammalian PRMTs for regulators of IFN-β expression and found that PRMT6 inhibits the antiviral innate immune response. Viral infection also upregulated PRMT6 protein levels. We generated PRMT6-deficient mice and found that they exhibited enhanced antiviral innate immunity. PRMT6 deficiency promoted the TBK1-IRF3 interaction and subsequently enhanced IRF3 activation and type-I interferon production. Mechanistically, viral infection enhanced the binding of PRMT6 to IRF3 and inhibited the interaction between IRF3 and TBK1; this mechanism was independent of PRMT6 methyltransferase activity. Thus, PRMT6 inhibits antiviral innate immunity by sequestering IRF3, thereby blocking TBK1-IRF3 signaling. Our work demonstrates a methyltransferase-independent role for PRMTs. It also identifies a negative regulator of the antiviral immune response, which may protect the host from the damaging effects of an overactive immune system and/or be exploited by viruses to escape immune detection.
Keywords: IFN; IRF3; PRMT6; TBK1; antiviral innate immunity.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





Similar articles
-
Grass carp PRMT6 negatively regulates innate immunity by inhibiting the TBK1/IRF3 binding and cutting down IRF3 phosphorylation level.Dev Comp Immunol. 2022 Apr;129:104351. doi: 10.1016/j.dci.2022.104351. Epub 2022 Jan 13. Dev Comp Immunol. 2022. PMID: 35033573
-
HSV-1 reactivation results in post-herpetic neuralgia by upregulating Prmt6 and inhibiting cGAS-STING.Brain. 2024 Jul 5;147(7):2552-2565. doi: 10.1093/brain/awae053. Brain. 2024. PMID: 38366606 Free PMC article.
-
Black carp PRMT6 inhibits TBK1-IRF3/7 signaling during the antiviral innate immune activation.Fish Shellfish Immunol. 2019 Oct;93:108-115. doi: 10.1016/j.fsi.2019.07.044. Epub 2019 Jul 18. Fish Shellfish Immunol. 2019. PMID: 31326582
-
TANK-binding kinase 1 as a novel therapeutic target for viral diseases.Expert Opin Ther Targets. 2019 May;23(5):437-446. doi: 10.1080/14728222.2019.1601702. Epub 2019 Apr 10. Expert Opin Ther Targets. 2019. PMID: 30932713 Review.
-
Negative regulation of TBK1-mediated antiviral immunity.FEBS Lett. 2013 Mar 18;587(6):542-8. doi: 10.1016/j.febslet.2013.01.052. Epub 2013 Feb 8. FEBS Lett. 2013. PMID: 23395611 Free PMC article. Review.
Cited by
-
PRMT6 Epigenetically Drives Metabolic Switch from Fatty Acid Oxidation toward Glycolysis and Promotes Osteoclast Differentiation During Osteoporosis.Adv Sci (Weinh). 2024 Oct;11(40):e2403177. doi: 10.1002/advs.202403177. Epub 2024 Aug 9. Adv Sci (Weinh). 2024. PMID: 39120025 Free PMC article.
-
Chicken PRMT3 facilitates IBDV replication.Poult Sci. 2024 Sep;103(9):103989. doi: 10.1016/j.psj.2024.103989. Epub 2024 Jun 17. Poult Sci. 2024. PMID: 38981362 Free PMC article.
-
The Role of Protein Methyltransferases in Immunity.Molecules. 2024 Jan 11;29(2):360. doi: 10.3390/molecules29020360. Molecules. 2024. PMID: 38257273 Free PMC article. Review.
-
The emerging roles of protein arginine methyltransferases in antiviral innate immune signaling pathways.Front Microbiol. 2023 Dec 5;14:1322929. doi: 10.3389/fmicb.2023.1322929. eCollection 2023. Front Microbiol. 2023. PMID: 38116532 Free PMC article. Review.
-
Protein arginine methylation in viral infection and antiviral immunity.Int J Biol Sci. 2023 Oct 24;19(16):5292-5318. doi: 10.7150/ijbs.89498. eCollection 2023. Int J Biol Sci. 2023. PMID: 37928266 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

