Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein
- PMID: 29967538
- PMCID: PMC6045459
- DOI: 10.1038/s41594-018-0083-z
Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein
Abstract
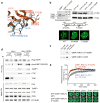
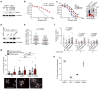
Dynamic protein interaction networks such as DNA double-strand break (DSB) signaling are modulated by post-translational modifications. The DNA repair factor 53BP1 is a rare example of a protein whose post-translational modification-binding function can be switched on and off. 53BP1 is recruited to DSBs by recognizing histone lysine methylation within chromatin, an activity directly inhibited by the 53BP1-binding protein TIRR. X-ray crystal structures of TIRR and a designer protein bound to 53BP1 now reveal a unique regulatory mechanism in which an intricate binding area centered on an essential TIRR arginine residue blocks the methylated-chromatin-binding surface of 53BP1. A 53BP1 separation-of-function mutation that abolishes TIRR-mediated regulation in cells renders 53BP1 hyperactive in response to DSBs, highlighting the key inhibitory function of TIRR. This 53BP1 inhibition is relieved by TIRR-interacting RNA molecules, providing proof-of-principle of RNA-triggered 53BP1 recruitment to DSBs.
Conflict of interest statement
The authors declare no competing interests.
Figures
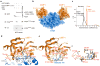


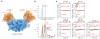

Similar articles
-
Molecular basis for the inhibition of the methyl-lysine binding function of 53BP1 by TIRR.Nat Commun. 2018 Jul 12;9(1):2689. doi: 10.1038/s41467-018-05174-9. Nat Commun. 2018. PMID: 30002377 Free PMC article.
-
TIRR regulates 53BP1 by masking its histone methyl-lysine binding function.Nature. 2017 Mar 9;543(7644):211-216. doi: 10.1038/nature21358. Epub 2017 Feb 27. Nature. 2017. PMID: 28241136 Free PMC article.
-
Structural basis for recognition of 53BP1 tandem Tudor domain by TIRR.Nat Commun. 2018 May 29;9(1):2123. doi: 10.1038/s41467-018-04557-2. Nat Commun. 2018. PMID: 29844495 Free PMC article.
-
TIRR: a potential front runner in HDR race-hypotheses and perspectives.Mol Biol Rep. 2020 Mar;47(3):2371-2379. doi: 10.1007/s11033-020-05285-x. Epub 2020 Feb 8. Mol Biol Rep. 2020. PMID: 32036573 Review.
-
Regulation of DNA double-strand break repair pathway choice: a new focus on 53BP1.J Zhejiang Univ Sci B. 2021 Jan 15;22(1):38-46. doi: 10.1631/jzus.B2000306. J Zhejiang Univ Sci B. 2021. PMID: 33448186 Free PMC article. Review.
Cited by
-
Ubiquitin Phosphorylation at Thr12 Modulates the DNA Damage Response.Mol Cell. 2020 Nov 5;80(3):423-436.e9. doi: 10.1016/j.molcel.2020.09.017. Epub 2020 Oct 5. Mol Cell. 2020. PMID: 33022275 Free PMC article.
-
53BP1 deficiency leads to hyperrecombination using break-induced replication (BIR).Nat Commun. 2024 Oct 5;15(1):8648. doi: 10.1038/s41467-024-52916-z. Nat Commun. 2024. PMID: 39368985 Free PMC article.
-
Biosensor for deconvolution of individual cell fate in response to ion beam irradiation.Cell Rep Methods. 2022 Feb 17;2(2):100169. doi: 10.1016/j.crmeth.2022.100169. eCollection 2022 Feb 28. Cell Rep Methods. 2022. PMID: 35474967 Free PMC article.
-
ATM-phosphorylated SPOP contributes to 53BP1 exclusion from chromatin during DNA replication.Sci Adv. 2021 Jun 18;7(25):eabd9208. doi: 10.1126/sciadv.abd9208. Print 2021 Jun. Sci Adv. 2021. PMID: 34144977 Free PMC article.
-
Mass spectrometry-based protein‒protein interaction techniques and their applications in studies of DNA damage repair.J Zhejiang Univ Sci B. 2021 Jan 15;22(1):1-20. doi: 10.1631/jzus.B2000356. J Zhejiang Univ Sci B. 2021. PMID: 33448183 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

