The multifaceted contributions of mitochondria to cellular metabolism
- PMID: 29950572
- PMCID: PMC6541229
- DOI: 10.1038/s41556-018-0124-1
The multifaceted contributions of mitochondria to cellular metabolism
Abstract
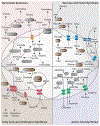
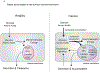
Although classically appreciated for their role as the powerhouse of the cell, the metabolic functions of mitochondria reach far beyond bioenergetics. In this Review, we discuss how mitochondria catabolize nutrients for energy, generate biosynthetic precursors for macromolecules, compartmentalize metabolites for the maintenance of redox homeostasis and function as hubs for metabolic waste management. We address the importance of these roles in both normal physiology and in disease.
Conflict of interest statement
Figures

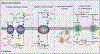

Comment in
-
Focusing on mitochondrial form and function.Nat Cell Biol. 2018 Jul;20(7):735. doi: 10.1038/s41556-018-0139-7. Nat Cell Biol. 2018. PMID: 29950569 No abstract available.
Similar articles
-
Metabolic Reprogramming in Modulating T Cell Reactive Oxygen Species Generation and Antioxidant Capacity.Front Immunol. 2018 May 16;9:1075. doi: 10.3389/fimmu.2018.01075. eCollection 2018. Front Immunol. 2018. PMID: 29868027 Free PMC article. Review.
-
Peroxisomal regulation of redox homeostasis and adipocyte metabolism.Redox Biol. 2019 Jun;24:101167. doi: 10.1016/j.redox.2019.101167. Epub 2019 Mar 14. Redox Biol. 2019. PMID: 30921635 Free PMC article. Review.
-
Reactive Oxygen Species and Mitochondrial Homeostasis as Regulators of Stem Cell Fate and Function.Antioxid Redox Signal. 2018 Jul 10;29(2):149-168. doi: 10.1089/ars.2017.7273. Epub 2017 Oct 26. Antioxid Redox Signal. 2018. PMID: 28708000 Review.
-
Redox imbalance and metabolic defects in the context of Alzheimer disease.FEBS Lett. 2024 Sep;598(17):2047-2066. doi: 10.1002/1873-3468.14840. Epub 2024 Mar 12. FEBS Lett. 2024. PMID: 38472147 Review.
-
Mitochondrial redox biology and homeostasis in plants.Trends Plant Sci. 2007 Mar;12(3):125-34. doi: 10.1016/j.tplants.2007.01.005. Epub 2007 Feb 12. Trends Plant Sci. 2007. PMID: 17293156 Review.
Cited by
-
Mitochondrial quality control in alcohol-associated liver disease.Hepatol Commun. 2024 Oct 24;8(11):e0534. doi: 10.1097/HC9.0000000000000534. eCollection 2024 Nov 1. Hepatol Commun. 2024. PMID: 39445886 Free PMC article. Review.
-
Unveiling the link: exploring muscle oxygen saturation in fibromyalgia and its implications for symptomatology and therapeutic strategies.Med Gas Res. 2025 Mar 1;15(1):58-72. doi: 10.4103/mgr.MEDGASRES-D-24-00013. Epub 2024 Apr 21. Med Gas Res. 2025. PMID: 39436169 Free PMC article. Review.
-
The role of reactive oxygen species in gastric cancer.Cancer Biol Med. 2024 Jul 9;21(9):740-53. doi: 10.20892/j.issn.2095-3941.2024.0182. Cancer Biol Med. 2024. PMID: 38982978 Free PMC article. Review.
-
Compensatory activity of the PC-ME1 metabolic axis underlies differential sensitivity to mitochondrial complex I inhibition.Nat Commun. 2024 Oct 7;15(1):8682. doi: 10.1038/s41467-024-52968-1. Nat Commun. 2024. PMID: 39375345 Free PMC article.
-
Chlamydia trachomatis Alters Mitochondrial Protein Composition and Secretes Effector Proteins That Target Mitochondria.mSphere. 2022 Dec 21;7(6):e0042322. doi: 10.1128/msphere.00423-22. Epub 2022 Oct 26. mSphere. 2022. PMID: 36286535 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

