Peritumoral monocytes induce cancer cell autophagy to facilitate the progression of human hepatocellular carcinoma
- PMID: 29940792
- PMCID: PMC6103724
- DOI: 10.1080/15548627.2018.1474994
Peritumoral monocytes induce cancer cell autophagy to facilitate the progression of human hepatocellular carcinoma
Abstract
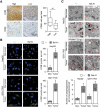
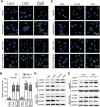
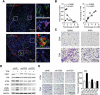
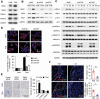
Macroautophagy/autophagy is an important catabolic process mediating cellular homeostasis and plays critical roles in cancer development. Whereas autophagy has been widely studied in various pathological models, little is known about the distribution, clinical significance and regulatory mechanism of this process in human hepatocellular carcinoma (HCC). In the present study, we found that tumor tissues exhibited significantly increased levels of autophagy compared with non-tumor tissues, and cancer cells with higher levels of autophagy were predominantly enriched in the invading edge regions of human HCC. Increased MAP1LC3B/LC3B expression in the invading edge regions was significantly correlated with a higher density of closely located monocytes, and TNF and IL1B derived from tumor-activated monocytes synergistically induced cancer cell autophagy in the invading edge regions of HCC. Monocyte-elicited autophagy induced the epithelial-mesenchymal transition (EMT) of cancer cells and promoted tumor metastasis by activating the NFKB-SNAI1 signaling pathway. Moreover, the increase of LC3B+ cancer cells in the invading edge areas was associated with high mortality and reduced survival of patients with HCC. These findings indicated that cancer cell autophagy is regulated by a collaborative interaction between tumor and immune cell components in distinct HCC microenvironments, thus allowing the inflammatory monocytes to be rerouted in a tumor-promoting direction.
Keywords: Autophagosome; epithelial-mesenchymal transition; invading edge; nuclear factor kappa-light-chain-enhancer of activated B cells (NFKB); tumor microenvironment.
Figures


Similar articles
-
FoxM1 overexpression promotes epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma.World J Gastroenterol. 2015 Jan 7;21(1):196-213. doi: 10.3748/wjg.v21.i1.196. World J Gastroenterol. 2015. PMID: 25574092 Free PMC article.
-
Mesencephalic Astrocyte-Derived Neurotrophic Factor Inhibits Liver Cancer Through Small Ubiquitin-Related Modifier (SUMO)ylation-Related Suppression of NF-κB/Snail Signaling Pathway and Epithelial-Mesenchymal Transition.Hepatology. 2020 Apr;71(4):1262-1278. doi: 10.1002/hep.30917. Epub 2020 Jan 26. Hepatology. 2020. PMID: 31469428 Free PMC article.
-
Apigenin inhibits NF-κB and snail signaling, EMT and metastasis in human hepatocellular carcinoma.Oncotarget. 2016 Jul 5;7(27):41421-41431. doi: 10.18632/oncotarget.9404. Oncotarget. 2016. PMID: 27203387 Free PMC article.
-
Modulation of the Autophagy-lysosomal Pathway in Hepatocellular Carcinoma Using Small Molecules.Molecules. 2020 Mar 30;25(7):1580. doi: 10.3390/molecules25071580. Molecules. 2020. PMID: 32235537 Free PMC article. Review.
-
Multipotent mesenchymal stromal cells play critical roles in hepatocellular carcinoma initiation, progression and therapy.Mol Cancer. 2018 Dec 28;17(1):178. doi: 10.1186/s12943-018-0926-6. Mol Cancer. 2018. PMID: 30593276 Free PMC article. Review.
Cited by
-
Intrahepatic inflammatory IgA+PD-L1high monocytes in hepatocellular carcinoma development and immunotherapy.J Immunother Cancer. 2022 May;10(5):e003618. doi: 10.1136/jitc-2021-003618. J Immunother Cancer. 2022. PMID: 35577505 Free PMC article.
-
Cellular based immunotherapy for primary liver cancer.J Exp Clin Cancer Res. 2021 Aug 9;40(1):250. doi: 10.1186/s13046-021-02030-5. J Exp Clin Cancer Res. 2021. PMID: 34372912 Free PMC article. Review.
-
The Autophagic Route of E-Cadherin and Cell Adhesion Molecules in Cancer Progression.Cancers (Basel). 2021 Dec 16;13(24):6328. doi: 10.3390/cancers13246328. Cancers (Basel). 2021. PMID: 34944948 Free PMC article. Review.
-
MiR-449a regulates the cell migration and invasion of human non-small cell lung carcinoma by targeting ADAM10.Onco Targets Ther. 2019 May 16;12:3829-3838. doi: 10.2147/OTT.S190282. eCollection 2019. Onco Targets Ther. 2019. PMID: 31190882 Free PMC article.
-
Predicting the recurrence of hepatocellular carcinoma (≤ 5 cm) after resection surgery with promising risk factors: habitat fraction of tumor and its peritumoral micro-environment.Radiol Med. 2023 Oct;128(10):1181-1191. doi: 10.1007/s11547-023-01695-6. Epub 2023 Aug 19. Radiol Med. 2023. PMID: 37597123
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
