Tie-Break: Host and Retrotransposons Play tRNA
- PMID: 29934075
- PMCID: PMC6520983
- DOI: 10.1016/j.tcb.2018.05.006
Tie-Break: Host and Retrotransposons Play tRNA
Abstract
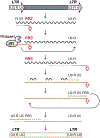
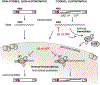
tRNA fragments (tRFs) are a class of small, regulatory RNAs with diverse functions. 3'-Derived tRFs perfectly match long terminal repeat (LTR)-retroelements which use the 3'-end of tRNAs to prime reverse transcription. Recent work has shown that tRFs target LTR-retroviruses and -transposons for the RNA interference (RNAi) pathway and also inhibit mobility by blocking reverse transcription. The highly conserved tRNA primer binding site (PBS) in LTR-retroelements is a unique target for 3'-tRFs to recognize and block abundant but diverse LTR-retrotransposons that become transcriptionally active during epigenetic reprogramming in development and disease. 3'-tRFs are processed from full-length tRNAs under so far unknown conditions and potentially protect many cell types. tRFs appear to be an ancient link between RNAi, transposons, and genome stability.
Keywords: LTR-retrotransposon; primer binding site; retrovirus; small RNA; tRF; tRNA.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Figures
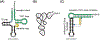
Similar articles
-
LTR-Retrotransposon Control by tRNA-Derived Small RNAs.Cell. 2017 Jun 29;170(1):61-71.e11. doi: 10.1016/j.cell.2017.06.013. Cell. 2017. PMID: 28666125 Free PMC article.
-
Endogenous Retroviruses Walk a Fine Line between Priming and Silencing.Viruses. 2020 Jul 23;12(8):792. doi: 10.3390/v12080792. Viruses. 2020. PMID: 32718022 Free PMC article. Review.
-
tRNA-derived small RNAs: New players in genome protection against retrotransposons.RNA Biol. 2018 Feb 1;15(2):170-175. doi: 10.1080/15476286.2017.1403000. Epub 2017 Dec 21. RNA Biol. 2018. PMID: 29120263 Free PMC article. Review.
-
tRNAs as primer of reverse transcriptases.Biochimie. 1995;77(1-2):113-24. doi: 10.1016/0300-9084(96)88114-4. Biochimie. 1995. PMID: 7541250 Review.
-
A novel mechanism of self-primed reverse transcription defines a new family of retroelements.Mol Cell Biol. 1995 Jun;15(6):3310-7. doi: 10.1128/MCB.15.6.3310. Mol Cell Biol. 1995. PMID: 7760826 Free PMC article.
Cited by
-
Noncanonical Roles of tRNAs: tRNA Fragments and Beyond.Annu Rev Genet. 2020 Nov 23;54:47-69. doi: 10.1146/annurev-genet-022620-101840. Epub 2020 Aug 25. Annu Rev Genet. 2020. PMID: 32841070 Free PMC article. Review.
-
Diversification of piRNAs expressed in PGCs and somatic cells during embryonic gonadal development.RNA Biol. 2020 Sep;17(9):1309-1323. doi: 10.1080/15476286.2020.1757908. Epub 2020 May 6. RNA Biol. 2020. PMID: 32375541 Free PMC article.
-
Strain-Specific Epigenetic Regulation of Endogenous Retroviruses: The Role of Trans-Acting Modifiers.Viruses. 2020 Jul 27;12(8):810. doi: 10.3390/v12080810. Viruses. 2020. PMID: 32727076 Free PMC article. Review.
-
tRNA Fragments Populations Analysis in Mutants Affecting tRNAs Processing and tRNA Methylation.Front Genet. 2020 Oct 9;11:518949. doi: 10.3389/fgene.2020.518949. eCollection 2020. Front Genet. 2020. PMID: 33193603 Free PMC article.
-
Idiosyncrasies of Viral Noncoding RNAs Provide Insights into Host Cell Biology.Annu Rev Virol. 2019 Sep 29;6(1):297-317. doi: 10.1146/annurev-virology-092818-015811. Epub 2019 Apr 30. Annu Rev Virol. 2019. PMID: 31039329 Free PMC article. Review.
References
-
- Peaston AE et al. (2004) Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell 7 (4), 597–606. - PubMed
-
- Faulkner GJ et al. (2009) The regulated retrotransposon transcriptome of mammalian cells. Nat Genet 41 (5), 563–71. - PubMed
-
- Siomi MC et al. (2011) PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 12 (4), 246–58. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

