Nucleolin facilitates nuclear retention of an ultraconserved region containing TRA2β4 and accelerates colon cancer cell growth
- PMID: 29928487
- PMCID: PMC6003563
- DOI: 10.18632/oncotarget.25510
Nucleolin facilitates nuclear retention of an ultraconserved region containing TRA2β4 and accelerates colon cancer cell growth
Abstract
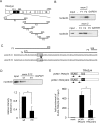
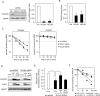
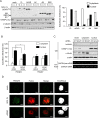
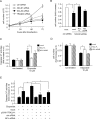
Transcribed-ultraconserved regions (T-UCRs), which contain conserved sequences with 100% identity across human, rat and mouse species, are a novel category of functional RNAs. The human transformer 2β gene (TRA2B) encodes a UCR that spans exon 2 (276 bp) and its neighboring introns. Among five spliced RNA variants (TRA2β1-5) transcribed from the TRA2B gene, only TRA2β4 contains the conserved exon 2. TRA2β4 is overexpressed in colon cancer cells and accelerates cell growth by blocking the transcription of CDKN1A. However, the mechanisms underlying the overexpression of TRA2β4 in colon cancer cells are unknown. Using biotinylated RNA pull-down assays followed by liquid chromatography-mass spectrometric analysis, we identified nucleolin as a TRA2β4-binding protein. Knockdown of nucleolin reduced the nuclear retention of TRA2β4 and accelerated its degradation in the cytoplasm, whereas nucleolin overexpression increased TRA2β4 levels and its mitogenic activity. Nucleolin directly bound to TRA2β4 exon 2 via the glycine/arginine-rich (GAR) domain. Overexpression of GAR-deficient nucleolin failed to increase TRA2β4 expression and growth of colon cancer cells. RNA fluorescence in situ hybridization showed that TRA2β4 co-localized with nucleolin in nuclei but not with the mutant lacking GAR. Our results suggest that specific interactions between nucleolin and UCR-containing TRA2β4 may be associated with abnormal growth of colon cancer cells.
Keywords: TRA2β4; cell growth; colon cancer; nucleolin; transcribed-UCR.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare that they have no conflict of interest.
Figures


Similar articles
-
HnRNPA1 interacts with G-quadruplex in the TRA2B promoter and stimulates its transcription in human colon cancer cells.Sci Rep. 2019 Jul 16;9(1):10276. doi: 10.1038/s41598-019-46659-x. Sci Rep. 2019. PMID: 31311954 Free PMC article.
-
Overexpression of the transcribed ultraconserved region Uc.138 accelerates colon cancer progression.Sci Rep. 2021 Apr 21;11(1):8667. doi: 10.1038/s41598-021-88123-9. Sci Rep. 2021. PMID: 33883665 Free PMC article.
-
Ultraconserved region-containing Transformer 2β4 controls senescence of colon cancer cells.Oncogenesis. 2016 Apr 4;5(4):e213. doi: 10.1038/oncsis.2016.18. Oncogenesis. 2016. PMID: 27043659 Free PMC article.
-
Highlighting transcribed ultraconserved regions in human diseases.Wiley Interdiscip Rev RNA. 2020 Mar;11(2):e1567. doi: 10.1002/wrna.1567. Epub 2019 Sep 5. Wiley Interdiscip Rev RNA. 2020. PMID: 31489780 Review.
-
Understanding the Genomic Ultraconservations: T-UCRs and Cancer.Int Rev Cell Mol Biol. 2017;333:159-172. doi: 10.1016/bs.ircmb.2017.04.004. Epub 2017 Jun 7. Int Rev Cell Mol Biol. 2017. PMID: 28729024 Review.
Cited by
-
Yeast Nucleolin Nsr1 Impedes Replication and Elevates Genome Instability at an Actively Transcribed Guanine-Rich G4 DNA-Forming Sequence.Genetics. 2020 Dec;216(4):1023-1037. doi: 10.1534/genetics.120.303736. Epub 2020 Oct 26. Genetics. 2020. PMID: 33106247 Free PMC article.
-
Cell surface nucleolin is a novel ADAMTS5 receptor mediating endothelial cell apoptosis.Cell Death Dis. 2022 Feb 23;13(2):172. doi: 10.1038/s41419-022-04618-x. Cell Death Dis. 2022. PMID: 35197459 Free PMC article.
-
HnRNPA1 interacts with G-quadruplex in the TRA2B promoter and stimulates its transcription in human colon cancer cells.Sci Rep. 2019 Jul 16;9(1):10276. doi: 10.1038/s41598-019-46659-x. Sci Rep. 2019. PMID: 31311954 Free PMC article.
-
Overexpression of the transcribed ultraconserved region Uc.138 accelerates colon cancer progression.Sci Rep. 2021 Apr 21;11(1):8667. doi: 10.1038/s41598-021-88123-9. Sci Rep. 2021. PMID: 33883665 Free PMC article.
-
Targeted splicing therapy: new strategies for colorectal cancer.Front Oncol. 2023 Aug 17;13:1222932. doi: 10.3389/fonc.2023.1222932. eCollection 2023. Front Oncol. 2023. PMID: 37664052 Free PMC article. Review.
References
-
- Dauwalder B, Amaya-Manzanares F, Mattox W. A human homologue of the Drosophila sex determination factor transformer-2 has conserved splicing regulatory functions. Proc Natl Acad Sci U S A. 1996;93:9004–9. https://doi.org/10.1073/pnas.93.17.9004. - DOI - PMC - PubMed
-
- Tsuda K, Someya T, Kuwasako K, Takahashi M, He F, Unzai S, Inoue M, Harada T, Watanabe S, Terada T, Kobayashi N, Shirouzu M, Kigawa T, et al. Structural basis for the dual RNA-recognition modes of human Tra2-β RRM. Nucleic Acids Res. 2011;39:1538–53. https://doi.org/10.1093/nar/gkq854. - DOI - PMC - PubMed
-
- Watermann DO, Tang Y, Zur Hausen A, Jäger M, Stamm S, Stickeler E. Splicing factor Tra2-β1 is specifically induced in breast cancer and regulates alternative splicing of the CD44 gene. Cancer Res. 2006;66:4774–80. https://doi.org/10.1158/0008-5472.CAN-04-3294. - DOI - PubMed
-
- Gabriel B, Zur Hausen A, Bouda J, Boudova L, Koprivova M, Hirschfeld M, Jager M, Stickeler E. Significance of nuclear hTra2-beta1 expression in cervical cancer. Acta Obstet Gynecol Scand. 2009;88:216–21. https://doi.org/10.1080/00016340802503021. - DOI - PubMed
-
- Kuwano Y, Nishida K, Kajita K, Satake Y, Akaike Y, Fujita K, Kano S, Masuda K, Rokutan K. Transformer 2β and miR-204 regulate apoptosis through competitive binding to 3′ UTR of BCL2 mRNA. Cell Death Differ. 2015;22:815–25. https://doi.org/10.1038/cdd.2014.176. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

