The E3 ligases Itch and WWP2 cooperate to limit TH2 differentiation by enhancing signaling through the TCR
- PMID: 29925997
- PMCID: PMC6348860
- DOI: 10.1038/s41590-018-0137-8
The E3 ligases Itch and WWP2 cooperate to limit TH2 differentiation by enhancing signaling through the TCR
Abstract
The mechanisms by which the sensitivity of naive CD4+ T cells to stimulation by the cognate antigen via the T cell antigen receptor (TCR) determines their differentiation into distinct helper T cell subsets remain elusive. Here we demonstrate functional collaboration of the ubiquitin E3 ligases Itch and WWP2 in regulating the strength of the TCR signal. Mice lacking both Itch and WWP2 in T cells showed spontaneous autoimmunity and lung inflammation. CD4+ T cells deficient in Itch and WWP2 exhibited hypo-responsiveness to TCR stimulation and a bias toward differentiation into the TH2 subset of helper T cells. Itch and WWP2 formed a complex and cooperated to enhance TCR-proximal signaling by catalyzing the conjugation of atypical ubiquitin chains to the phosphatase SHP-1 and reducing the association of SHP-1 with the tyrosine kinase Lck. These findings indicate that targeted ubiquitination regulates the strength of the TCR signal and differentiation toward the TH2 lineage.
Figures

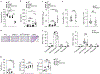


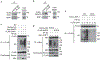

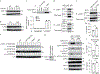
Similar articles
-
K33-linked polyubiquitination of T cell receptor-zeta regulates proteolysis-independent T cell signaling.Immunity. 2010 Jul 23;33(1):60-70. doi: 10.1016/j.immuni.2010.07.002. Immunity. 2010. PMID: 20637659 Free PMC article.
-
TAOK3 Regulates Canonical TCR Signaling by Preventing Early SHP-1-Mediated Inactivation of LCK.J Immunol. 2018 Dec 1;201(11):3431-3442. doi: 10.4049/jimmunol.1800284. Epub 2018 Oct 29. J Immunol. 2018. PMID: 30373850
-
Immune regulation by protein ubiquitination: roles of the E3 ligases VHL and Itch.Protein Cell. 2019 Jun;10(6):395-404. doi: 10.1007/s13238-018-0586-8. Epub 2018 Nov 9. Protein Cell. 2019. PMID: 30413999 Free PMC article. Review.
-
Deficient expression of p56(lck) in Th2 cells leads to partial TCR signaling and a dysregulation in lymphokine mRNA levels.J Immunol. 1996 Dec 1;157(11):4751-61. J Immunol. 1996. PMID: 8943376
-
TAOK3, a Regulator of LCK-SHP-1 Crosstalk during TCR Signaling.Crit Rev Immunol. 2019;39(1):59-81. doi: 10.1615/CritRevImmunol.2019030480. Crit Rev Immunol. 2019. PMID: 31679194 Review.
Cited by
-
A novel human fetal lung-derived alveolar organoid model reveals mechanisms of surfactant protein C maturation relevant to interstitial lung disease.bioRxiv [Preprint]. 2023 Sep 4:2023.08.30.555522. doi: 10.1101/2023.08.30.555522. bioRxiv. 2023. Update in: EMBO J. 2025 Feb;44(3):639-664. doi: 10.1038/s44318-024-00328-6. PMID: 37693487 Free PMC article. Updated. Preprint.
-
Hectd3 promotes pathogenic Th17 lineage through Stat3 activation and Malt1 signaling in neuroinflammation.Nat Commun. 2019 Feb 11;10(1):701. doi: 10.1038/s41467-019-08605-3. Nat Commun. 2019. PMID: 30741923 Free PMC article.
-
Potential of E3 Ubiquitin Ligases in Cancer Immunity: Opportunities and Challenges.Cells. 2021 Nov 25;10(12):3309. doi: 10.3390/cells10123309. Cells. 2021. PMID: 34943817 Free PMC article. Review.
-
The Ubiquitin-Proteasome System in Immune Cells.Biomolecules. 2021 Jan 5;11(1):60. doi: 10.3390/biom11010060. Biomolecules. 2021. PMID: 33466553 Free PMC article. Review.
-
Beyond the Cell Surface: Targeting Intracellular Negative Regulators to Enhance T cell Anti-Tumor Activity.Int J Mol Sci. 2019 Nov 20;20(23):5821. doi: 10.3390/ijms20235821. Int J Mol Sci. 2019. PMID: 31756921 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

