Independent evolution of functionally exchangeable mitochondrial outer membrane import complexes
- PMID: 29923829
- PMCID: PMC6010339
- DOI: 10.7554/eLife.34488
Independent evolution of functionally exchangeable mitochondrial outer membrane import complexes
Abstract
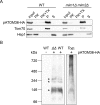
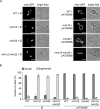
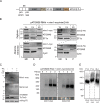
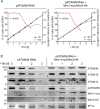
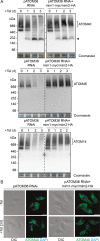
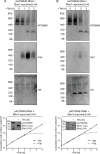
Assembly and/or insertion of a subset of mitochondrial outer membrane (MOM) proteins, including subunits of the main MOM translocase, require the fungi-specific Mim1/Mim2 complex. So far it was unclear which proteins accomplish this task in other eukaryotes. Here, we show by reciprocal complementation that the MOM protein pATOM36 of trypanosomes is a functional analogue of yeast Mim1/Mim2 complex, even though these proteins show neither sequence nor topological similarity. Expression of pATOM36 rescues almost all growth, mitochondrial biogenesis, and morphology defects in yeast cells lacking Mim1 and/or Mim2. Conversely, co-expression of Mim1 and Mim2 restores the assembly and/or insertion defects of MOM proteins in trypanosomes ablated for pATOM36. Mim1/Mim2 and pATOM36 form native-like complexes when heterologously expressed, indicating that additional proteins are not part of these structures. Our findings indicate that Mim1/Mim2 and pATOM36 are the products of convergent evolution and arose only after the ancestors of fungi and trypanosomatids diverged.
Keywords: MIM complex; S. cerevisiae; biochemistry; biogenesis; chemical biology; mitochondria; outer membrane; trypanosome.
© 2018, Vitali et al.
Conflict of interest statement
DV, SK, AK, KD, AS, DR No competing interests declared
Figures







Comment in
-
Shaping the import system of mitochondria.Elife. 2018 Jun 20;7:e38209. doi: 10.7554/eLife.38209. Elife. 2018. PMID: 29923828 Free PMC article.
Similar articles
-
A crucial role for Mim2 in the biogenesis of mitochondrial outer membrane proteins.J Cell Sci. 2012 Jul 15;125(Pt 14):3464-73. doi: 10.1242/jcs.103804. Epub 2012 Mar 30. J Cell Sci. 2012. PMID: 22467864
-
An essential novel component of the noncanonical mitochondrial outer membrane protein import system of trypanosomatids.Mol Biol Cell. 2012 Sep;23(17):3420-8. doi: 10.1091/mbc.E12-02-0107. Epub 2012 Jul 11. Mol Biol Cell. 2012. PMID: 22787278 Free PMC article.
-
Outer membrane protein functions as integrator of protein import and DNA inheritance in mitochondria.Proc Natl Acad Sci U S A. 2016 Aug 2;113(31):E4467-75. doi: 10.1073/pnas.1605497113. Epub 2016 Jul 19. Proc Natl Acad Sci U S A. 2016. PMID: 27436903 Free PMC article.
-
The enigmatic role of Mim1 in mitochondrial biogenesis.Eur J Cell Biol. 2010 Feb-Mar;89(2-3):212-5. doi: 10.1016/j.ejcb.2009.11.002. Epub 2009 Nov 26. Eur J Cell Biol. 2010. PMID: 19944477 Review.
-
Mitochondrial protein import in trypanosomes: Expect the unexpected.Traffic. 2017 Feb;18(2):96-109. doi: 10.1111/tra.12463. Epub 2017 Jan 17. Traffic. 2017. PMID: 27976830 Review.
Cited by
-
Crosstalk between Mitochondrial Protein Import and Lipids.Int J Mol Sci. 2022 May 9;23(9):5274. doi: 10.3390/ijms23095274. Int J Mol Sci. 2022. PMID: 35563660 Free PMC article. Review.
-
Protein control of membrane and organelle dynamics: Insights from the divergent eukaryote Toxoplasma gondii.Curr Opin Cell Biol. 2022 Jun;76:102085. doi: 10.1016/j.ceb.2022.102085. Epub 2022 May 12. Curr Opin Cell Biol. 2022. PMID: 35569259 Free PMC article. Review.
-
Protein Import into the Endosymbiotic Organelles of Apicomplexan Parasites.Genes (Basel). 2018 Aug 14;9(8):412. doi: 10.3390/genes9080412. Genes (Basel). 2018. PMID: 30110980 Free PMC article. Review.
-
p166 links membrane and intramitochondrial modules of the trypanosomal tripartite attachment complex.PLoS Pathog. 2022 Jun 16;18(6):e1010207. doi: 10.1371/journal.ppat.1010207. eCollection 2022 Jun. PLoS Pathog. 2022. PMID: 35709300 Free PMC article.
-
Targeting and Insertion of Membrane Proteins in Mitochondria.Front Cell Dev Biol. 2021 Dec 24;9:803205. doi: 10.3389/fcell.2021.803205. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 35004695 Free PMC article. Review.
References
-
- Becker T, Pfannschmidt S, Guiard B, Stojanovski D, Milenkovic D, Kutik S, Pfanner N, Meisinger C, Wiedemann N. Biogenesis of the mitochondrial TOM complex: mim1 promotes insertion and assembly of signal-anchored receptors. The Journal of Biological Chemistry. 2008a;283:120–127. doi: 10.1074/jbc.M706997200. - DOI - PubMed
-
- Becker T, Wenz LS, Krüger V, Lehmann W, Müller JM, Goroncy L, Zufall N, Lithgow T, Guiard B, Chacinska A, Wagner R, Meisinger C, Pfanner N. The mitochondrial import protein Mim1 promotes biogenesis of multispanning outer membrane proteins. The Journal of Cell Biology. 2011;194:387–395. doi: 10.1083/jcb.201102044. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

