Targeting Transcription Factors for Cancer Treatment
- PMID: 29921764
- PMCID: PMC6100431
- DOI: 10.3390/molecules23061479
Targeting Transcription Factors for Cancer Treatment
Abstract
Transcription factors are involved in a large number of human diseases such as cancers for which they account for about 20% of all oncogenes identified so far. For long time, with the exception of ligand-inducible nuclear receptors, transcription factors were considered as “undruggable” targets. Advances knowledge of these transcription factors, in terms of structure, function (expression, degradation, interaction with co-factors and other proteins) and the dynamics of their mode of binding to DNA has changed this postulate and paved the way for new therapies targeted against transcription factors. Here, we discuss various ways to target transcription factors in cancer models: by modulating their expression or degradation, by blocking protein/protein interactions, by targeting the transcription factor itself to prevent its DNA binding either through a binding pocket or at the DNA-interacting site, some of these inhibitors being currently used or evaluated for cancer treatment. Such different targeting of transcription factors by small molecules is facilitated by modern chemistry developing a wide variety of original molecules designed to specifically abort transcription factor and by an increased knowledge of their pathological implication through the use of new technologies in order to make it possible to improve therapeutic control of transcription factor oncogenic functions.
Keywords: DNA binding; inhibitors; oncogenes; protein/DNA interaction; protein/protein interaction; transcription factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
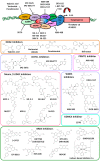
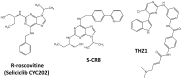
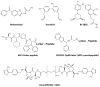
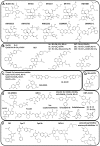
Similar articles
-
Targeting transcription factors by small compounds--Current strategies and future implications.Biochem Pharmacol. 2016 May 1;107:1-13. doi: 10.1016/j.bcp.2015.12.006. Epub 2015 Dec 11. Biochem Pharmacol. 2016. PMID: 26686579 Review.
-
Targeting Transcription Factor Binding to DNA by Competing with DNA Binders as an Approach for Controlling Gene Expression.Curr Top Med Chem. 2015;15(14):1323-58. doi: 10.2174/1568026615666150413154713. Curr Top Med Chem. 2015. PMID: 25866275 Review.
-
Targeting General Transcriptional Machinery as a Therapeutic Strategy for Adult T-Cell Leukemia.Molecules. 2018 May 2;23(5):1057. doi: 10.3390/molecules23051057. Molecules. 2018. PMID: 29724031 Free PMC article. Review.
-
Exploring Proteomic Drug Targets, Therapeutic Strategies and Protein - Protein Interactions in Cancer: Mechanistic View.Curr Cancer Drug Targets. 2019;19(6):430-448. doi: 10.2174/1568009618666180803104631. Curr Cancer Drug Targets. 2019. PMID: 30073927 Review.
-
Inhibition of BET bromodomains as a therapeutic strategy for cancer drug discovery.Oncotarget. 2015 Mar 20;6(8):5501-16. doi: 10.18632/oncotarget.3551. Oncotarget. 2015. PMID: 25849938 Free PMC article. Review.
Cited by
-
Emerging role of pioneer transcription factors in targeted ERα positive breast cancer.Explor Target Antitumor Ther. 2021;2(1):26-35. doi: 10.37349/etat.2021.00031. Epub 2021 Feb 28. Explor Target Antitumor Ther. 2021. PMID: 36046086 Free PMC article. Review.
-
The transcription factor TBP promotes hepatocellular carcinoma progression by activating AKT3.Am J Cancer Res. 2023 Nov 15;13(11):5656-5666. eCollection 2023. Am J Cancer Res. 2023. PMID: 38058816 Free PMC article.
-
Indirect Mechanisms of Transcription Factor-Mediated Gene Regulation during Cell Fate Changes.Adv Genet (Hoboken). 2022 Nov 9;3(4):2200015. doi: 10.1002/ggn2.202200015. eCollection 2022 Dec. Adv Genet (Hoboken). 2022. PMID: 36911290 Free PMC article.
-
Tumor suppressive BTB/POZ zinc-finger protein ZBTB28 inhibits oncogenic BCL6/ZBTB27 signaling to maintain p53 transcription in multiple carcinogenesis.Theranostics. 2019 Oct 18;9(26):8182-8195. doi: 10.7150/thno.34983. eCollection 2019. Theranostics. 2019. PMID: 31754389 Free PMC article.
-
IRF2 is a master regulator of human keratinocyte stem cell fate.Nat Commun. 2019 Oct 14;10(1):4676. doi: 10.1038/s41467-019-12559-x. Nat Commun. 2019. PMID: 31611556 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

