Computational design of orthogonal membrane receptor-effector switches for rewiring signaling pathways
- PMID: 29915030
- PMCID: PMC6142221
- DOI: 10.1073/pnas.1718489115
Computational design of orthogonal membrane receptor-effector switches for rewiring signaling pathways
Abstract
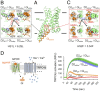
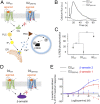
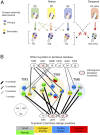
Membrane receptors regulate numerous intracellular functions. However, the molecular underpinnings remain poorly understood because most receptors initiate multiple signaling pathways through distinct interaction interfaces that are structurally uncharacterized. We present an integrated computational and experimental approach to model and rationally engineer membrane receptor-intracellular protein systems signaling with novel pathway selectivity. We targeted the dopamine D2 receptor (D2), a G-protein-coupled receptor (GPCR), which primarily signals through Gi, but triggers also the Gq and beta-arrestin pathways. Using this approach, we designed orthogonal D2-Gi complexes, which coupled with high specificity and triggered exclusively the Gi-dependent signaling pathway. We also engineered an orthogonal chimeric D2-Gs/i complex that rewired D2 signaling from a Gi-mediated inhibitory into a Gs-dependent activating pathway. Reinterpreting the evolutionary history of GPCRs in light of the designed proteins, we uncovered an unforeseen hierarchical code of GPCR-G-protein coupling selectivity determinants. The results demonstrate that membrane receptor-cytosolic protein systems can be rationally engineered to regulate mammalian cellular functions. The method should prove useful for creating orthogonal molecular switches that redirect signals at the cell surface for cell-engineering applications.
Keywords: G-protein–coupled receptor; cell signaling; membrane protein; protein binding; protein design.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Selective reconstitution of human D4 dopamine receptor variants with Gi alpha subtypes.Biochemistry. 2000 Apr 4;39(13):3734-44. doi: 10.1021/bi992354c. Biochemistry. 2000. PMID: 10736173
-
Evaluating cellular impedance assays for detection of GPCR pleiotropic signaling and functional selectivity.J Biomol Screen. 2009 Mar;14(3):246-55. doi: 10.1177/1087057108330115. Epub 2009 Feb 11. J Biomol Screen. 2009. PMID: 19211780
-
GHSR-D2R heteromerization modulates dopamine signaling through an effect on G protein conformation.Proc Natl Acad Sci U S A. 2018 Apr 24;115(17):4501-4506. doi: 10.1073/pnas.1712725115. Epub 2018 Apr 9. Proc Natl Acad Sci U S A. 2018. PMID: 29632174 Free PMC article.
-
Gs/Gi Regulation of Bone Cell Differentiation: Review and Insights from Engineered Receptors.Horm Metab Res. 2016 Nov;48(11):689-699. doi: 10.1055/s-0042-116156. Epub 2016 Sep 19. Horm Metab Res. 2016. PMID: 27643449 Review.
-
G protein pathways.Science. 2002 May 31;296(5573):1636-9. doi: 10.1126/science.1071550. Science. 2002. PMID: 12040175 Review.
Cited by
-
Protein Structure Prediction and Design in a Biologically Realistic Implicit Membrane.Biophys J. 2020 Apr 21;118(8):2042-2055. doi: 10.1016/j.bpj.2020.03.006. Epub 2020 Mar 14. Biophys J. 2020. PMID: 32224301 Free PMC article.
-
Reprogramming G protein coupled receptor structure and function.Curr Opin Struct Biol. 2018 Aug;51:187-194. doi: 10.1016/j.sbi.2018.07.008. Epub 2018 Jul 25. Curr Opin Struct Biol. 2018. PMID: 30055347 Free PMC article. Review.
-
Engineering signalling pathways in mammalian cells.Nat Biomed Eng. 2024 Dec;8(12):1523-1539. doi: 10.1038/s41551-024-01237-z. Epub 2024 Sep 5. Nat Biomed Eng. 2024. PMID: 39237709 Review.
-
Membrane Protein Engineering with Rosetta.Methods Mol Biol. 2021;2315:43-57. doi: 10.1007/978-1-0716-1468-6_3. Methods Mol Biol. 2021. PMID: 34302669 Free PMC article.
-
Computational design of G Protein-Coupled Receptor allosteric signal transductions.Nat Chem Biol. 2020 Jan;16(1):77-86. doi: 10.1038/s41589-019-0407-2. Epub 2019 Dec 2. Nat Chem Biol. 2020. PMID: 31792443
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

