Transposable elements generate regulatory novelty in a tissue-specific fashion
- PMID: 29914366
- PMCID: PMC6006921
- DOI: 10.1186/s12864-018-4850-3
Transposable elements generate regulatory novelty in a tissue-specific fashion
Abstract
Background: Transposable elements (TE) are an important source of evolutionary novelty in gene regulation. However, the mechanisms by which TEs contribute to gene expression are largely uncharacterized.
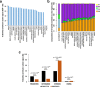
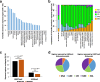
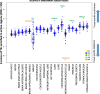
Results: Here, we leverage Roadmap and GTEx data to investigate the association of TEs with active and repressed chromatin in 24 tissues. We find 112 human TE families enriched in active regions of the genome across tissues. Short Interspersed Nuclear Elements (SINEs) and DNA transposons are the most frequently enriched classes, while Long Terminal Repeat Retrotransposons (LTRs) are often enriched in a tissue-specific manner. We report across-tissue variability in TE enrichment in active regions. Genes with consistent expression across tissues are less likely to be associated with TE insertions. TE presence in repressed regions similarly follows tissue-specific patterns. Moreover, different TE classes correlate with different repressive marks: LTRs and Long Interspersed Nuclear Elements (LINEs) are overrepresented in regions marked by H3K9me3, while the other TEs are more likely to overlap regions with H3K27me3. Young TEs are typically enriched in repressed regions and depleted in active regions. We detect multiple instances of TEs that are enriched in tissue-specific active regulatory regions. Such TEs contain binding sites for transcription factors that are master regulators for the given tissue. These TEs are enriched in intronic enhancers, and their tissue-specific enrichment correlates with tissue-specific variations in the expression of the nearest genes.
Conclusions: We provide an integrated overview of the contribution of TEs to human gene regulation. Expanding previous analyses, we demonstrate that TEs can potentially contribute to the turnover of regulatory sequences in a tissue-specific fashion.
Keywords: Gene regulation; Tissue-specific; Transcription factors; Transposons.
Conflict of interest statement
Ethics approval and consent to participate
N/A
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


Similar articles
-
Transposable Element Exaptation into Regulatory Regions Is Rare, Influenced by Evolutionary Age, and Subject to Pleiotropic Constraints.Mol Biol Evol. 2017 Nov 1;34(11):2856-2869. doi: 10.1093/molbev/msx219. Mol Biol Evol. 2017. PMID: 28961735 Free PMC article.
-
Specific subfamilies of transposable elements contribute to different domains of T lymphocyte enhancers.Proc Natl Acad Sci U S A. 2020 Apr 7;117(14):7905-7916. doi: 10.1073/pnas.1912008117. Epub 2020 Mar 19. Proc Natl Acad Sci U S A. 2020. PMID: 32193341 Free PMC article.
-
Genome-Wide Analysis of the Association of Transposable Elements with Gene Regulation Suggests that Alu Elements Have the Largest Overall Regulatory Impact.J Comput Biol. 2018 Jun;25(6):551-562. doi: 10.1089/cmb.2017.0228. Epub 2018 Apr 30. J Comput Biol. 2018. PMID: 29708779
-
Transposable element influences on gene expression in plants.Biochim Biophys Acta Gene Regul Mech. 2017 Jan;1860(1):157-165. doi: 10.1016/j.bbagrm.2016.05.010. Epub 2016 May 25. Biochim Biophys Acta Gene Regul Mech. 2017. PMID: 27235540 Review.
-
Origin of a substantial fraction of human regulatory sequences from transposable elements.Trends Genet. 2003 Feb;19(2):68-72. doi: 10.1016/s0168-9525(02)00006-9. Trends Genet. 2003. PMID: 12547512 Review.
Cited by
-
The Transposable Element Environment of Human Genes Differs According to Their Duplication Status and Essentiality.Genome Biol Evol. 2021 May 7;13(5):evab062. doi: 10.1093/gbe/evab062. Genome Biol Evol. 2021. PMID: 33973013 Free PMC article.
-
Tissue-specific usage of transposable element-derived promoters in mouse development.Genome Biol. 2020 Sep 28;21(1):255. doi: 10.1186/s13059-020-02164-3. Genome Biol. 2020. PMID: 32988383 Free PMC article.
-
AGO2 silences mobile transposons in the nucleus of quiescent cells.Nat Struct Mol Biol. 2023 Dec;30(12):1985-1995. doi: 10.1038/s41594-023-01151-z. Epub 2023 Nov 20. Nat Struct Mol Biol. 2023. PMID: 37985687
-
Roles of Transposable Elements in the Different Layers of Gene Expression Regulation.Int J Mol Sci. 2019 Nov 15;20(22):5755. doi: 10.3390/ijms20225755. Int J Mol Sci. 2019. PMID: 31731828 Free PMC article. Review.
-
Genomic features underlie the co-option of SVA transposons as cis-regulatory elements in human pluripotent stem cells.PLoS Genet. 2022 Jun 15;18(6):e1010225. doi: 10.1371/journal.pgen.1010225. eCollection 2022 Jun. PLoS Genet. 2022. PMID: 35704668 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

