EMI1 switches from being a substrate to an inhibitor of APC/CCDH1 to start the cell cycle
- PMID: 29875408
- PMCID: PMC6035873
- DOI: 10.1038/s41586-018-0199-7
EMI1 switches from being a substrate to an inhibitor of APC/CCDH1 to start the cell cycle
Abstract
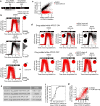
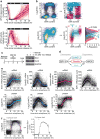
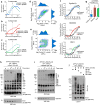
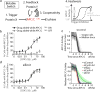
Mammalian cells integrate mitogen and stress signalling before the end of G1 phase to determine whether or not they enter the cell cycle1-4. Before cells can replicate their DNA in S phase, they have to activate cyclin-dependent kinases (CDKs), induce an E2F transcription program and inactivate the anaphase-promoting complex (APC/CCDH1, also known as the cyclosome), which is an E3 ubiquitin ligase that contains the co-activator CDH1 (also known as FZR, encoded by FZR1). It was recently shown that stress can return cells to quiescence after CDK2 activation and E2F induction but not after inactivation of APC/CCDH1, which suggests that APC/CCDH1 inactivation is the point of no return for cell-cycle entry 3 . Rapid inactivation of APC/CCDH1 requires early mitotic inhibitor 1 (EMI1)3,5, but the molecular mechanism that controls this cell-cycle commitment step is unknown. Here we show using human cell models that cell-cycle commitment is mediated by an EMI1-APC/CCDH1 dual-negative feedback switch, in which EMI1 is both a substrate and an inhibitor of APC/CCDH1. The inactivation switch triggers a transition between a state with low EMI1 levels and high APC/CCDH1 activity during G1 and a state with high EMI1 levels and low APC/CCDH1 activity during S and G2. Cell-based analysis, in vitro reconstitution and modelling data show that the underlying dual-negative feedback is bistable and represents a robust irreversible switch. Our study suggests that mammalian cells commit to the cell cycle by increasing CDK2 activity and EMI1 mRNA expression to trigger a one-way APC/CCDH1 inactivation switch that is mediated by EMI1 transitioning from acting as a substrate of APC/CCDH1 to being an inhibitor of APC/CCDH1.
Conflict of interest statement
M.R. is a co-founder and consultant to Nurix, a biotech company in the ubiquitin space.
Figures








Similar articles
-
Irreversible APC(Cdh1) Inactivation Underlies the Point of No Return for Cell-Cycle Entry.Cell. 2016 Jun 30;166(1):167-80. doi: 10.1016/j.cell.2016.05.077. Cell. 2016. PMID: 27368103 Free PMC article.
-
Examining the mechanistic relationship of APC/CCDH1 and its interphase inhibitor EMI1.Protein Sci. 2022 Jun;31(6):e4324. doi: 10.1002/pro.4324. Protein Sci. 2022. PMID: 35634770 Free PMC article.
-
Cyclin A2-cyclin-dependent kinase 2 cooperates with the PLK1-SCFbeta-TrCP1-EMI1-anaphase-promoting complex/cyclosome axis to promote genome reduplication in the absence of mitosis.Mol Cell Biol. 2009 Dec;29(24):6500-14. doi: 10.1128/MCB.00669-09. Epub 2009 Oct 12. Mol Cell Biol. 2009. PMID: 19822658 Free PMC article.
-
Defining the role of Emi1 in the DNA replication-segregation cycle.Chromosoma. 2008 Aug;117(4):333-8. doi: 10.1007/s00412-008-0152-x. Epub 2008 Mar 4. Chromosoma. 2008. PMID: 18317792 Review.
-
Association of Hsp90 with p53 and Fizzy related homolog (Fzr) synchronizing Anaphase Promoting Complex (APC/C): An unexplored ally towards oncogenic pathway.Biochim Biophys Acta Rev Cancer. 2023 May;1878(3):188883. doi: 10.1016/j.bbcan.2023.188883. Epub 2023 Mar 25. Biochim Biophys Acta Rev Cancer. 2023. PMID: 36972769 Review.
Cited by
-
Control of DNA Replication Initiation by Ubiquitin.Cells. 2018 Sep 20;7(10):146. doi: 10.3390/cells7100146. Cells. 2018. PMID: 30241373 Free PMC article. Review.
-
Induction of Premature Cell Senescence Stimulated by High Doses of Antioxidants Is Mediated by Endoplasmic Reticulum Stress.Int J Mol Sci. 2021 Oct 31;22(21):11851. doi: 10.3390/ijms222111851. Int J Mol Sci. 2021. PMID: 34769282 Free PMC article.
-
Cell cycle inertia underlies a bifurcation in cell fates after DNA damage.Sci Adv. 2021 Jan 13;7(3):eabe3882. doi: 10.1126/sciadv.abe3882. Print 2021 Jan. Sci Adv. 2021. PMID: 33523889 Free PMC article.
-
The role of Anaphase Promoting Complex activation, inhibition and substrates in cancer development and progression.Aging (Albany NY). 2020 Aug 15;12(15):15818-15855. doi: 10.18632/aging.103792. Epub 2020 Aug 15. Aging (Albany NY). 2020. PMID: 32805721 Free PMC article. Review.
-
APC/C ubiquitin ligase: Functions and mechanisms in tumorigenesis.Semin Cancer Biol. 2020 Dec;67(Pt 2):80-91. doi: 10.1016/j.semcancer.2020.03.001. Epub 2020 Mar 9. Semin Cancer Biol. 2020. PMID: 32165320 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

