NF-κB-Dependent Lymphoid Enhancer Co-option Promotes Renal Carcinoma Metastasis
- PMID: 29875134
- PMCID: PMC6031301
- DOI: 10.1158/2159-8290.CD-17-1211
NF-κB-Dependent Lymphoid Enhancer Co-option Promotes Renal Carcinoma Metastasis
Abstract
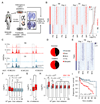
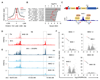
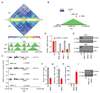
Metastases, the spread of cancer cells to distant organs, cause the majority of cancer-related deaths. Few metastasis-specific driver mutations have been identified, suggesting aberrant gene regulation as a source of metastatic traits. However, how metastatic gene expression programs arise is poorly understood. Here, using human-derived metastasis models of renal cancer, we identify transcriptional enhancers that promote metastatic carcinoma progression. Specific enhancers and enhancer clusters are activated in metastatic cancer cell populations, and the associated gene expression patterns are predictive of poor patient outcome in clinical samples. We find that the renal cancer metastasis-associated enhancer complement consists of multiple coactivated tissue-specific enhancer modules. Specifically, we identify and functionally characterize a coregulatory enhancer cluster, activated by the renal cancer driver HIF2A and an NF-κB-driven lymphoid element, as a mediator of metastasis in vivo We conclude that oncogenic pathways can acquire metastatic phenotypes through cross-lineage co-option of physiologic epigenetic enhancer states.Significance: Renal cancer is associated with significant mortality due to metastasis. We show that in metastatic renal cancer, functionally important metastasis genes are activated via co-option of gene regulatory enhancer modules from distant developmental lineages, thus providing clues to the origins of metastatic cancer. Cancer Discov; 8(7); 850-65. ©2018 AACR.This article is highlighted in the In This Issue feature, p. 781.
©2018 American Association for Cancer Research.
Conflict of interest statement
Figures


Similar articles
-
VHL Deficiency Drives Enhancer Activation of Oncogenes in Clear Cell Renal Cell Carcinoma.Cancer Discov. 2017 Nov;7(11):1284-1305. doi: 10.1158/2159-8290.CD-17-0375. Epub 2017 Sep 11. Cancer Discov. 2017. PMID: 28893800
-
Downregulated METTL14 accumulates BPTF that reinforces super-enhancers and distal lung metastasis via glycolytic reprogramming in renal cell carcinoma.Theranostics. 2021 Jan 26;11(8):3676-3693. doi: 10.7150/thno.55424. eCollection 2021. Theranostics. 2021. PMID: 33664855 Free PMC article.
-
Identification of activated enhancers and linked transcription factors in breast, prostate, and kidney tumors by tracing enhancer networks using epigenetic traits.Epigenetics Chromatin. 2016 Nov 9;9:50. doi: 10.1186/s13072-016-0102-4. eCollection 2016. Epigenetics Chromatin. 2016. PMID: 27833659 Free PMC article.
-
The emerging role of nuclear factor kappa B in renal cell carcinoma.Int J Biochem Cell Biol. 2011 Nov;43(11):1537-49. doi: 10.1016/j.biocel.2011.08.003. Epub 2011 Aug 12. Int J Biochem Cell Biol. 2011. PMID: 21854869 Review.
-
Enhancer reprogramming in tumor progression: a new route towards cancer cell plasticity.Cell Mol Life Sci. 2018 Jul;75(14):2537-2555. doi: 10.1007/s00018-018-2820-1. Epub 2018 Apr 24. Cell Mol Life Sci. 2018. PMID: 29691590 Free PMC article. Review.
Cited by
-
Endogenous HIF2A reporter systems for high-throughput functional screening.Sci Rep. 2018 Aug 13;8(1):12063. doi: 10.1038/s41598-018-30499-2. Sci Rep. 2018. PMID: 30104738 Free PMC article.
-
Genomic control of metastasis.Br J Cancer. 2021 Jan;124(1):3-12. doi: 10.1038/s41416-020-01127-6. Epub 2020 Nov 4. Br J Cancer. 2021. PMID: 33144692 Free PMC article. Review.
-
SETD2 loss perturbs the kidney cancer epigenetic landscape to promote metastasis and engenders actionable dependencies on histone chaperone complexes.Nat Cancer. 2022 Feb;3(2):188-202. doi: 10.1038/s43018-021-00316-3. Epub 2022 Feb 3. Nat Cancer. 2022. PMID: 35115713 Free PMC article.
-
Editorial: Multidisciplinary Approaches in Exploring Cancer Heterogeneity, TME and Therapy Resistance: Perspectives for Systems Medicine.Front Cell Dev Biol. 2022 Feb 3;10:842596. doi: 10.3389/fcell.2022.842596. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35198561 Free PMC article. No abstract available.
-
Multi-Omics Profiling to Assess Signaling Changes upon VHL Restoration and Identify Putative VHL Substrates in Clear Cell Renal Cell Carcinoma Cell Lines.Cells. 2022 Jan 29;11(3):472. doi: 10.3390/cells11030472. Cells. 2022. PMID: 35159281 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

