Complete Genome Characterization of the 2017 Dengue Outbreak in Xishuangbanna, a Border City of China, Burma and Laos
- PMID: 29868504
- PMCID: PMC5951998
- DOI: 10.3389/fcimb.2018.00148
Complete Genome Characterization of the 2017 Dengue Outbreak in Xishuangbanna, a Border City of China, Burma and Laos
Abstract
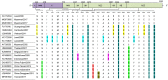
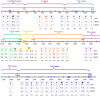
A dengue outbreak abruptly occurred at the border of China, Myanmar, and Laos in June 2017. By November 3rd 2017, 1184 infected individuals were confirmed as NS1-positivein Xishuangbanna, a city located at the border. To verify the causative agent, complete genome information was obtained through PCR and sequencing based on the viral RNAs extracted from patient samples. Phylogenetic trees were constructed by the maximum likelihood method (MEGA 6.0). Nucleotide and amino acid substitutions were analyzed by BioEdit, followed by RNA secondary structure prediction of untranslated regions (UTRs) and protein secondary structure prediction in coding sequences (CDSs). Strains YN2, YN17741, and YN176272 were isolated from local residents. Stains MY21 and MY22 were isolated from Burmese travelers. The complete genome sequences of the five isolates were 10,735 nucleotides in length. Phylogenetic analysis classified all five isolates as genotype I of DENV-1, while isolates of local residents and Burmese travelers belonged to different branches. The three locally isolates were most similar to the Dongguan strain in 2011, and the other two isolates from Burmese travelers were most similar to the Laos strain in 2008. Twenty-four amino acid substitutions were important in eight evolutionary tree branches. Comparison with DENV-1SS revealed 658 base substitutions in the local isolates, except for two mutations exclusive to YN17741, resulting in 87 synonymous mutations. Compared with the local isolates, 52 amino acid mutations occurred in the CDS of two isolates from Burmese travelers. Comparing MY21 with MY22, 17 amino acid mutations were observed, all these mutations occurred in the CDS of non-structured proteins (two in NS1, 10 in NS2, two in NS3, three in NS5). Secondary structure prediction revealed 46 changes in the potential nucleotide and protein binding sites of the CDSs in local isolates. RNA secondary structure prediction also showed base changes in the 3'UTR of local isolates, leading to two significant changes in the RNA secondary structure. To our knowledge, this study is the first complete genome analysis of isolates from the 2017 dengue outbreak that occurred at the border areas of China, Burma, and Laos.
Keywords: RNA secondary structure; complete genome; dengue outbreak; phylogenetic trees; protein secondary structure.
Figures


Similar articles
-
Complete genome analysis of dengue virus type 3 isolated from the 2013 dengue outbreak in Yunnan, China.Virus Res. 2017 Jun 15;238:164-170. doi: 10.1016/j.virusres.2017.06.015. Epub 2017 Jun 23. Virus Res. 2017. PMID: 28648850
-
Molecular Characterization of Dengue Virus Serotype 2 Cosmospolitan Genotype From 2015 Dengue Outbreak in Yunnan, China.Front Cell Infect Microbiol. 2018 Jun 27;8:219. doi: 10.3389/fcimb.2018.00219. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29998087 Free PMC article.
-
Molecular characterization of the viral structural gene of the first dengue virus type 1 outbreak in Xishuangbanna: A border area of China, Burma and Laos.Int J Infect Dis. 2019 Feb;79:152-161. doi: 10.1016/j.ijid.2018.11.370. Epub 2018 Dec 5. Int J Infect Dis. 2019. PMID: 30528395
-
Molecular characterization of an imported dengue virus serotype 4 isolate from Thailand.Arch Virol. 2018 Oct;163(10):2903-2906. doi: 10.1007/s00705-018-3906-7. Epub 2018 Jun 14. Arch Virol. 2018. PMID: 29948381 Review.
-
Functional RNA elements in the dengue virus genome.Viruses. 2011 Sep;3(9):1739-56. doi: 10.3390/v3091739. Epub 2011 Sep 15. Viruses. 2011. PMID: 21994804 Free PMC article. Review.
Cited by
-
Molecular characteristics of the VP1 region of enterovirus 71 strains in China.Gut Pathog. 2020 Aug 14;12:38. doi: 10.1186/s13099-020-00377-2. eCollection 2020. Gut Pathog. 2020. PMID: 32818043 Free PMC article.
-
FLAVi: An Enhanced Annotator for Viral Genomes of Flaviviridae.Viruses. 2020 Aug 14;12(8):892. doi: 10.3390/v12080892. Viruses. 2020. PMID: 32824044 Free PMC article.
-
Neutralizing antibodies from prior exposure to dengue virus negatively correlate with viremia on re-infection.Commun Med (Lond). 2023 Oct 19;3(1):148. doi: 10.1038/s43856-023-00378-7. Commun Med (Lond). 2023. PMID: 37857747 Free PMC article.
-
Retrospective investigation of the origin and epidemiology of the dengue outbreak in Yunnan, China from 2017 to 2018.Front Vet Sci. 2023 Apr 3;10:1137392. doi: 10.3389/fvets.2023.1137392. eCollection 2023. Front Vet Sci. 2023. PMID: 37124563 Free PMC article.
-
Genetic Characterization of Chikungunya Virus Among Febrile Dengue Fever-Like Patients in Xishuangbanna, Southwestern Part of China.Front Cell Infect Microbiol. 2022 Jun 27;12:914289. doi: 10.3389/fcimb.2022.914289. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35832380 Free PMC article.
References
-
- Añez G., Volkova E., Jiang Z., Heisey D. A. R., Chancey C., Fares R. C. G. (2017). Collaborative study to establish World Health Organization international reference reagents for dengue virus Types 1 to 4 RNA for use in nucleic acid testing. Transfusion 57, 1977–1987. 10.1111/trf.14130 - DOI - PubMed
-
- Butrapet S., Huang C. Y., Pierro D. J., Bhamarapravati N., Gubler D. J., Kinney R. M. (2000). Attenuation markers of a candidate dengue type 2 vaccine virus, strain 16681 (PDK-53), are defined by mutations in the 5' noncoding region and nonstructural proteins 1 and 3. J. Virol. 74, 3011–3019. 10.1128/JVI.74.7.3011-3019.2000 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

