Plant Viral Proteases: Beyond the Role of Peptide Cutters
- PMID: 29868107
- PMCID: PMC5967125
- DOI: 10.3389/fpls.2018.00666
Plant Viral Proteases: Beyond the Role of Peptide Cutters
Abstract
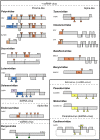
Almost half of known plant viral species rely on proteolytic cleavages as key co- and post-translational modifications throughout their infection cycle. Most of these viruses encode their own endopeptidases, proteases with high substrate specificity that internally cleave large polyprotein precursors for the release of functional sub-units. Processing of the polyprotein, however, is not an all-or-nothing process in which endopeptidases act as simple peptide cutters. On the contrary, spatial-temporal modulation of these polyprotein cleavage events is crucial for a successful viral infection. In this way, the processing of the polyprotein coordinates viral replication, assembly and movement, and has significant impact on pathogen fitness and virulence. In this mini-review, we give an overview of plant viral proteases emphasizing their importance during viral infections and the varied functionalities that result from their proteolytic activities.
Keywords: defense and counterdefense; host range; plant viruses; viral polyprotein; viral proteases; viral replication; virion formation.
Figures
Similar articles
-
Strawberry Mottle Virus (Family Secoviridae, Order Picornavirales) Encodes a Novel Glutamic Protease To Process the RNA2 Polyprotein at Two Cleavage Sites.J Virol. 2019 Feb 19;93(5):e01679-18. doi: 10.1128/JVI.01679-18. Print 2019 Mar 1. J Virol. 2019. PMID: 30541838 Free PMC article.
-
Proteolytic Processing of Plant Proteins by Potyvirus NIa Proteases.J Virol. 2022 Jan 26;96(2):e0144421. doi: 10.1128/JVI.01444-21. Epub 2021 Nov 10. J Virol. 2022. PMID: 34757836 Free PMC article.
-
Thrombin cleavage of the hepatitis E virus polyprotein at multiple conserved locations is required for genome replication.PLoS Pathog. 2023 Jul 21;19(7):e1011529. doi: 10.1371/journal.ppat.1011529. eCollection 2023 Jul. PLoS Pathog. 2023. PMID: 37478143 Free PMC article.
-
Expanding Repertoire of Plant Positive-Strand RNA Virus Proteases.Viruses. 2019 Jan 15;11(1):66. doi: 10.3390/v11010066. Viruses. 2019. PMID: 30650571 Free PMC article. Review.
-
Proteomics approaches for the identification of protease substrates during virus infection.Adv Virus Res. 2021;109:135-161. doi: 10.1016/bs.aivir.2021.03.003. Epub 2021 Apr 20. Adv Virus Res. 2021. PMID: 33934826 Review.
Cited by
-
Strawberry Mottle Virus (Family Secoviridae, Order Picornavirales) Encodes a Novel Glutamic Protease To Process the RNA2 Polyprotein at Two Cleavage Sites.J Virol. 2019 Feb 19;93(5):e01679-18. doi: 10.1128/JVI.01679-18. Print 2019 Mar 1. J Virol. 2019. PMID: 30541838 Free PMC article.
-
Proteolytic Processing of Plant Proteins by Potyvirus NIa Proteases.J Virol. 2022 Jan 26;96(2):e0144421. doi: 10.1128/JVI.01444-21. Epub 2021 Nov 10. J Virol. 2022. PMID: 34757836 Free PMC article.
-
Pepper Mottle Virus and Its Host Interactions: Current State of Knowledge.Viruses. 2021 Sep 25;13(10):1930. doi: 10.3390/v13101930. Viruses. 2021. PMID: 34696360 Free PMC article. Review.
-
Viral deubiquitinating proteases and the promising strategies of their inhibition.Virus Res. 2024 Jun;344:199368. doi: 10.1016/j.virusres.2024.199368. Epub 2024 Apr 11. Virus Res. 2024. PMID: 38588924 Free PMC article. Review.
-
On the cutting edge: protease-based methods for sensing and controlling cell biology.Nat Methods. 2020 Sep;17(9):885-896. doi: 10.1038/s41592-020-0891-z. Epub 2020 Jul 13. Nat Methods. 2020. PMID: 32661424 Review.
References
-
- Agranovsky A. A. (2016). “Closteroviruses: molecular biology, evolution and interactions with cells,” in eds Gaur R. K., Petrov N. M., Patil B. L., Stoyanova M. I. (Singapore: Springer; ).
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

