Stochastic Seeding Coupled with mRNA Self-Recruitment Generates Heterogeneous Drosophila Germ Granules
- PMID: 29861136
- PMCID: PMC6008217
- DOI: 10.1016/j.cub.2018.04.037
Stochastic Seeding Coupled with mRNA Self-Recruitment Generates Heterogeneous Drosophila Germ Granules
Abstract
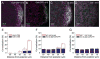
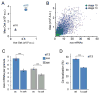
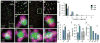
The formation of ribonucleoprotein assemblies called germ granules is a conserved feature of germline development. In Drosophila, germ granules form at the posterior of the oocyte in a specialized cytoplasm called the germ plasm, which specifies germline fate during embryogenesis. mRNAs, including nanos (nos) and polar granule component (pgc), that function in germline development are localized to the germ plasm through their incorporation into germ granules, which deliver them to the primordial germ cells. Germ granules are nucleated by Oskar (Osk) protein and contain varying combinations and quantities of their constituent mRNAs, which are organized as spatially distinct, multi-copy homotypic clusters. The process that gives rise to such heterogeneous yet organized granules remains unknown. Here, we show that individual nos and pgc transcripts can populate the same nascent granule, and these first transcripts then act as seeds, recruiting additional like transcripts to form homotypic clusters. Within a granule, homotypic clusters grow independently of each other but depend on the simultaneous acquisition of additional Osk. Although granules can contain multiple clusters of a particular mRNA, granule mRNA content is dominated by cluster size. These results suggest that the accumulation of mRNAs in the germ plasm is controlled by the mRNAs themselves through their ability to form homotypic clusters; thus, RNA self-association drives germ granule mRNA localization. We propose that a stochastic seeding and self-recruitment mechanism enables granules to simultaneously incorporate many different mRNAs while ensuring that each becomes enriched to a functional threshold.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures

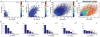

Similar articles
-
Distinct cis-acting elements mediate targeting and clustering of Drosophila polar granule mRNAs.Development. 2018 Nov 22;145(22):dev164657. doi: 10.1242/dev.164657. Development. 2018. PMID: 30333216 Free PMC article.
-
Germ Granule Evolution Provides Mechanistic Insight into Drosophila Germline Development.Mol Biol Evol. 2023 Aug 3;40(8):msad174. doi: 10.1093/molbev/msad174. Mol Biol Evol. 2023. PMID: 37527522 Free PMC article.
-
Compartmentalized oskar degradation in the germ plasm safeguards germline development.Elife. 2020 Jan 7;9:e49988. doi: 10.7554/eLife.49988. Elife. 2020. PMID: 31909715 Free PMC article.
-
Germ granules in Drosophila.Traffic. 2019 Sep;20(9):650-660. doi: 10.1111/tra.12674. Epub 2019 Jul 31. Traffic. 2019. PMID: 31218815 Free PMC article. Review.
-
The role of mitochondrial rRNAs and nanos protein in germline formation in Drosophila embryos.Zoolog Sci. 2005 Sep;22(9):943-54. doi: 10.2108/zsj.22.943. Zoolog Sci. 2005. PMID: 16219975 Review.
Cited by
-
Caenorhabditis elegans germ granules accumulate hundreds of low translation mRNAs with no systematic preference for germ cell fate regulators.Development. 2024 Jul 1;151(13):dev202575. doi: 10.1242/dev.202575. Epub 2024 Jul 10. Development. 2024. PMID: 38984542 Free PMC article.
-
Distinct cis-acting elements mediate targeting and clustering of Drosophila polar granule mRNAs.Development. 2018 Nov 22;145(22):dev164657. doi: 10.1242/dev.164657. Development. 2018. PMID: 30333216 Free PMC article.
-
L-bodies are RNA-protein condensates driving RNA localization in Xenopus oocytes.Mol Biol Cell. 2021 Dec 1;32(22):ar37. doi: 10.1091/mbc.E21-03-0146-T. Epub 2021 Oct 6. Mol Biol Cell. 2021. PMID: 34613784 Free PMC article.
-
Mutational analysis of the functional motifs of the DEAD-box RNA helicase Me31B/DDX6 in Drosophila germline development.FEBS Lett. 2023 Jul;597(14):1848-1867. doi: 10.1002/1873-3468.14668. Epub 2023 Jun 8. FEBS Lett. 2023. PMID: 37235728 Free PMC article.
-
RNA Granules: A View from the RNA Perspective.Molecules. 2020 Jul 8;25(14):3130. doi: 10.3390/molecules25143130. Molecules. 2020. PMID: 32650583 Free PMC article. Review.
References
-
- Kato Y, Nakamura A. Roles of cytoplasmic RNP granules in intracellular RNA localization and translational control in the Drosophila oocyte. Dev Growth Differ. 2012;54:19–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

