Integrated Phloem Sap mRNA and Protein Expression Analysis Reveals Phytoplasma-infection Responses in Mulberry
- PMID: 29848783
- PMCID: PMC6126391
- DOI: 10.1074/mcp.RA118.000670
Integrated Phloem Sap mRNA and Protein Expression Analysis Reveals Phytoplasma-infection Responses in Mulberry
Abstract
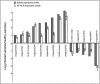
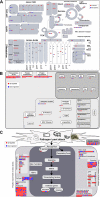
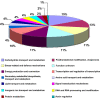
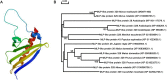
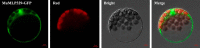
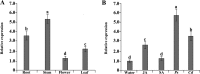
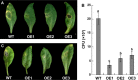
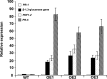
To gain insight into the response of mulberry to phytoplasma-infection, the expression profiles of mRNAs and proteins in mulberry phloem sap were examined. A total of 955 unigenes and 136 proteins were found to be differentially expressed between the healthy and infected phloem sap. These differentially expressed mRNAs and proteins are involved in signaling, hormone metabolism, stress responses, etc. Interestingly, we found that both the mRNA and protein levels of the major latex protein-like 329 (MuMLPL329) gene were increased in the infected phloem saps. Expression of the MuMLPL329 gene was induced by pathogen inoculation and was responsive to jasmonic acid. Ectopic expression of MuMLPL329 in Arabidopsis enhances transgenic plant resistance to Botrytis cinerea, Pseudomonas syringae pv tomato DC3000 (Pst. DC3000) and phytoplasma. Further analysis revealed that MuMLPL329 can enhance the expression of some defense genes and might be involved in altering flavonoid content resulting in increased resistance of plants to pathogen infection. Finally, the roles of the differentially expressed mRNAs and proteins and the potential molecular mechanisms of their changes were discussed. It was likely that the phytoplasma-responsive mRNAs and proteins in the phloem saps were involved in multiple pathways of mulberry responses to phytoplasma-infection, and their changes may be partially responsible for some symptoms in the phytoplasma infected plants.
© 2018 Gai et al.
Conflict of interest statement
Conflict of interest: We declare no conflict of interest.
Figures





Similar articles
-
MiRNA-seq-based profiles of miRNAs in mulberry phloem sap provide insight into the pathogenic mechanisms of mulberry yellow dwarf disease.Sci Rep. 2018 Jan 16;8(1):812. doi: 10.1038/s41598-018-19210-7. Sci Rep. 2018. PMID: 29339758 Free PMC article.
-
Metabolomic analysis reveals the potential metabolites and pathogenesis involved in mulberry yellow dwarf disease.Plant Cell Environ. 2014 Jun;37(6):1474-90. doi: 10.1111/pce.12255. Epub 2014 Jan 9. Plant Cell Environ. 2014. PMID: 24329897
-
Analysis of phytoplasma-responsive sRNAs provide insight into the pathogenic mechanisms of mulberry yellow dwarf disease.Sci Rep. 2014 Jun 20;4:5378. doi: 10.1038/srep05378. Sci Rep. 2014. PMID: 24946736 Free PMC article.
-
Comparative proteomic analysis provides new insights into mulberry dwarf responses in mulberry (Morus alba L.).Proteomics. 2009 Dec;9(23):5328-39. doi: 10.1002/pmic.200900012. Proteomics. 2009. PMID: 19834890
-
Heterologous expression of Chinese wild grapevine VqERFs in Arabidopsis thaliana enhance resistance to Pseudomonas syringae pv. tomato DC3000 and to Botrytis cinerea.Plant Sci. 2020 Apr;293:110421. doi: 10.1016/j.plantsci.2020.110421. Epub 2020 Jan 23. Plant Sci. 2020. PMID: 32081269
Cited by
-
A newly identified glycosyltransferase AsRCOM provides resistance to purple curl leaf disease in agave.BMC Genomics. 2023 Nov 7;24(1):669. doi: 10.1186/s12864-023-09700-y. BMC Genomics. 2023. PMID: 37936069 Free PMC article.
-
Plasmodesmata and their role in the regulation of phloem unloading during fruit development.Curr Opin Plant Biol. 2021 Dec;64:102145. doi: 10.1016/j.pbi.2021.102145. Epub 2021 Nov 23. Curr Opin Plant Biol. 2021. PMID: 34826657 Free PMC article. Review.
-
Transcriptome and DNA Methylome Reveal Insights Into Phytoplasma Infection Responses in Mulberry (Morus multicaulis Perr.).Front Plant Sci. 2021 Aug 3;12:697702. doi: 10.3389/fpls.2021.697702. eCollection 2021. Front Plant Sci. 2021. PMID: 34413866 Free PMC article.
-
Nitric oxide (NO) modulates low temperature-stress signaling via S-nitrosation, a NO PTM, inducing ethylene biosynthesis inhibition leading to enhanced post-harvest shelf-life of agricultural produce.Physiol Mol Biol Plants. 2023 Dec;29(12):2051-2065. doi: 10.1007/s12298-023-01371-z. Epub 2023 Nov 2. Physiol Mol Biol Plants. 2023. PMID: 38222283 Free PMC article. Review.
-
Major latex-like proteins show pH dependency in their binding to hydrophobic organic pollutants.J Pestic Sci. 2023 Aug 20;48(3):71-77. doi: 10.1584/jpestics.D23-014. J Pestic Sci. 2023. PMID: 37745171 Free PMC article.
References
-
- Kehr J., and Buhtz A. (2008) Long distance transport and movement of RNA through the phloem. J. Exp. Bot. 59, 85–92 - PubMed
-
- Crawford K. M., and Zambryski P. C. (1999) Phloem transport: are you chaperoned? Curr. Biol. 9, R281–R285 - PubMed
-
- Heo J. O., Roszak P., Furuta K. M., and Helariutta Y. (2014) Phloem development: Current knowledge and future perspectives. Am. J. Bot. 101, 1393–1402 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

