The HIV-1 Tat protein recruits a ubiquitin ligase to reorganize the 7SK snRNP for transcriptional activation
- PMID: 29845934
- PMCID: PMC5999396
- DOI: 10.7554/eLife.31879
The HIV-1 Tat protein recruits a ubiquitin ligase to reorganize the 7SK snRNP for transcriptional activation
Abstract
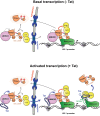
The HIV-1 Tat protein hijacks P-TEFb kinase to activate paused RNA polymerase II (RNAP II) at the viral promoter. Tat binds additional host factors, but it is unclear how they regulate RNAP II elongation. Here, we identify the cytoplasmic ubiquitin ligase UBE2O as critical for Tat transcriptional activity. Tat hijacks UBE2O to ubiquitinate the P-TEFb kinase inhibitor HEXIM1 of the 7SK snRNP, a fraction of which also resides in the cytoplasm bound to P-TEFb. HEXIM1 ubiquitination sequesters it in the cytoplasm and releases P-TEFb from the inhibitory 7SK complex. Free P-TEFb then becomes enriched in chromatin, a process that is also stimulated by treating cells with a CDK9 inhibitor. Finally, we demonstrate that UBE2O is critical for P-TEFb recruitment to the HIV-1 promoter. Together, the data support a unique model of elongation control where non-degradative ubiquitination of nuclear and cytoplasmic 7SK snRNP pools increases P-TEFb levels for transcriptional activation.
Keywords: 7SK snRNP; HIV-1; biochemistry; chemical biology; host-pathogen interactions; human; non-degradative ubiquitination; nuclear import; transcription elongation.
© 2018, Faust et al.
Conflict of interest statement
TF, YL, CB, GJ, AW, BJ, BN, NK, ID, AF No competing interests declared
Figures









Similar articles
-
Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat.PLoS Pathog. 2010 Oct 14;6(10):e1001152. doi: 10.1371/journal.ppat.1001152. PLoS Pathog. 2010. PMID: 20976203 Free PMC article.
-
The mechanism of release of P-TEFb and HEXIM1 from the 7SK snRNP by viral and cellular activators includes a conformational change in 7SK.PLoS One. 2010 Aug 23;5(8):e12335. doi: 10.1371/journal.pone.0012335. PLoS One. 2010. PMID: 20808803 Free PMC article.
-
A human immunodeficiency virus type 1 Tat-like arginine-rich RNA-binding domain is essential for HEXIM1 to inhibit RNA polymerase II transcription through 7SK snRNA-mediated inactivation of P-TEFb.Mol Cell Biol. 2004 Jun;24(12):5094-105. doi: 10.1128/MCB.24.12.5094-5105.2004. Mol Cell Biol. 2004. PMID: 15169877 Free PMC article.
-
New insights into the control of HIV-1 transcription: when Tat meets the 7SK snRNP and super elongation complex (SEC).J Neuroimmune Pharmacol. 2011 Jun;6(2):260-8. doi: 10.1007/s11481-011-9267-6. Epub 2011 Mar 1. J Neuroimmune Pharmacol. 2011. PMID: 21360054 Free PMC article. Review.
-
Cracking the control of RNA polymerase II elongation by 7SK snRNP and P-TEFb.Nucleic Acids Res. 2016 Sep 19;44(16):7527-39. doi: 10.1093/nar/gkw585. Epub 2016 Jul 1. Nucleic Acids Res. 2016. PMID: 27369380 Free PMC article. Review.
Cited by
-
MicroRNAs and long non-coding RNAs during transcriptional regulation and latency of HIV and HTLV.Retrovirology. 2024 Feb 29;21(1):5. doi: 10.1186/s12977-024-00637-y. Retrovirology. 2024. PMID: 38424561 Free PMC article. Review.
-
CDK9: a signaling hub for transcriptional control.Transcription. 2019 Apr;10(2):57-75. doi: 10.1080/21541264.2018.1523668. Epub 2018 Oct 11. Transcription. 2019. PMID: 30227759 Free PMC article. Review.
-
Roles of Farnesyl-Diphosphate Farnesyltransferase 1 in Tumour and Tumour Microenvironments.Cells. 2020 Oct 25;9(11):2352. doi: 10.3390/cells9112352. Cells. 2020. PMID: 33113804 Free PMC article. Review.
-
An alpha-herpesvirus employs host HEXIM1 to promote viral transcription.J Virol. 2024 Mar 19;98(3):e0139223. doi: 10.1128/jvi.01392-23. Epub 2024 Feb 16. J Virol. 2024. PMID: 38363111 Free PMC article.
-
A functional map of HIV-host interactions in primary human T cells.Nat Commun. 2022 Apr 1;13(1):1752. doi: 10.1038/s41467-022-29346-w. Nat Commun. 2022. PMID: 35365639 Free PMC article.
References
-
- Bartholomeeusen K, Xiang Y, Fujinaga K, Peterlin BM. Bromodomain and extra-terminal (BET) bromodomain inhibition activate transcription via transient release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein. Journal of Biological Chemistry. 2012;287:36609–36616. doi: 10.1074/jbc.M112.410746. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

