dbCAN2: a meta server for automated carbohydrate-active enzyme annotation
- PMID: 29771380
- PMCID: PMC6031026
- DOI: 10.1093/nar/gky418
dbCAN2: a meta server for automated carbohydrate-active enzyme annotation
Abstract
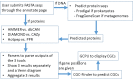
Complex carbohydrates of plants are the main food sources of animals and microbes, and serve as promising renewable feedstock for biofuel and biomaterial production. Carbohydrate active enzymes (CAZymes) are the most important enzymes for complex carbohydrate metabolism. With an increasing number of plant and plant-associated microbial genomes and metagenomes being sequenced, there is an urgent need of automatic tools for genomic data mining of CAZymes. We developed the dbCAN web server in 2012 to provide a public service for automated CAZyme annotation for newly sequenced genomes. Here, dbCAN2 (http://cys.bios.niu.edu/dbCAN2) is presented as an updated meta server, which integrates three state-of-the-art tools for CAZome (all CAZymes of a genome) annotation: (i) HMMER search against the dbCAN HMM (hidden Markov model) database; (ii) DIAMOND search against the CAZy pre-annotated CAZyme sequence database and (iii) Hotpep search against the conserved CAZyme short peptide database. Combining the three outputs and removing CAZymes found by only one tool can significantly improve the CAZome annotation accuracy. In addition, dbCAN2 now also accepts nucleotide sequence submission, and offers the service to predict physically linked CAZyme gene clusters (CGCs), which will be a very useful online tool for identifying putative polysaccharide utilization loci (PULs) in microbial genomes or metagenomes.
Figures

Similar articles
-
dbCAN3: automated carbohydrate-active enzyme and substrate annotation.Nucleic Acids Res. 2023 Jul 5;51(W1):W115-W121. doi: 10.1093/nar/gkad328. Nucleic Acids Res. 2023. PMID: 37125649 Free PMC article.
-
dbCAN-seq: a database of carbohydrate-active enzyme (CAZyme) sequence and annotation.Nucleic Acids Res. 2018 Jan 4;46(D1):D516-D521. doi: 10.1093/nar/gkx894. Nucleic Acids Res. 2018. PMID: 30053267 Free PMC article.
-
PlantCAZyme: a database for plant carbohydrate-active enzymes.Database (Oxford). 2014 Aug 14;2014:bau079. doi: 10.1093/database/bau079. Print 2014. Database (Oxford). 2014. PMID: 25125445 Free PMC article.
-
Combined whole cell wall analysis and streamlined in silico carbohydrate-active enzyme discovery to improve biocatalytic conversion of agricultural crop residues.Biotechnol Biofuels. 2021 Jan 9;14(1):16. doi: 10.1186/s13068-020-01869-8. Biotechnol Biofuels. 2021. PMID: 33422151 Free PMC article. Review.
-
Computational Glycobiology: Mechanistic Studies of Carbohydrate-Active Enzymes and Implication for Inhibitor Design.Adv Protein Chem Struct Biol. 2017;109:25-76. doi: 10.1016/bs.apcsb.2017.04.003. Epub 2017 May 19. Adv Protein Chem Struct Biol. 2017. PMID: 28683920 Review.
Cited by
-
Segatella clades adopt distinct roles within a single individual's gut.NPJ Biofilms Microbiomes. 2024 Oct 27;10(1):114. doi: 10.1038/s41522-024-00590-w. NPJ Biofilms Microbiomes. 2024. PMID: 39465298 Free PMC article.
-
The plant rhizosheath-root niche is an edaphic "mini-oasis" in hyperarid deserts with enhanced microbial competition.ISME Commun. 2022 Jun 3;2(1):47. doi: 10.1038/s43705-022-00130-7. ISME Commun. 2022. PMID: 37938683 Free PMC article.
-
Novel bacterial taxa in a minimal lignocellulolytic consortium and their potential for lignin and plastics transformation.ISME Commun. 2022 Sep 26;2(1):89. doi: 10.1038/s43705-022-00176-7. ISME Commun. 2022. PMID: 37938754 Free PMC article.
-
Bacillus altitudinis 1.4 genome analysis-functional annotation of probiotic properties and immunomodulatory activity.World J Microbiol Biotechnol. 2024 Aug 8;40(10):293. doi: 10.1007/s11274-024-04096-7. World J Microbiol Biotechnol. 2024. PMID: 39112831
-
Bagasse minority pathway expression: Real time study of GH2 β-mannosidases from bacteroidetes.PLoS One. 2021 Mar 17;16(3):e0247822. doi: 10.1371/journal.pone.0247822. eCollection 2021. PLoS One. 2021. PMID: 33730062 Free PMC article.
References
-
- Cockburn D.W., Koropatkin N.M.. Polysaccharide degradation by the intestinal microbiota and its influence on human health and disease. J. Mol. Biol. 2016; 428:3230–3252. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

