Discovery and Sequence Analysis of Four Deltacoronaviruses from Birds in the Middle East Reveal Interspecies Jumping with Recombination as a Potential Mechanism for Avian-to-Avian and Avian-to-Mammalian Transmission
- PMID: 29769348
- PMCID: PMC6052312
- DOI: 10.1128/JVI.00265-18
Discovery and Sequence Analysis of Four Deltacoronaviruses from Birds in the Middle East Reveal Interspecies Jumping with Recombination as a Potential Mechanism for Avian-to-Avian and Avian-to-Mammalian Transmission
Abstract
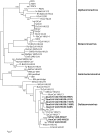
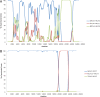
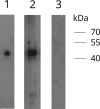
The emergence of Middle East respiratory syndrome showed once again that coronaviruses (CoVs) in animals are potential source for epidemics in humans. To explore the diversity of deltacoronaviruses in animals in the Middle East, we tested fecal samples from 1,356 mammals and birds in Dubai, The United Arab Emirates. Four novel deltacoronaviruses were detected from eight birds of four species by reverse transcription-PCR (RT-PCR): FalCoV UAE-HKU27 from a falcon, HouCoV UAE-HKU28 from a houbara bustard, PiCoV UAE-HKU29 from a pigeon, and QuaCoV UAE-HKU30 from five quails. Complete genome sequencing showed that FalCoV UAE-HKU27, HouCoV UAE-HKU28, and PiCoV UAE-HKU29 belong to the same CoV species, suggesting recent interspecies transmission between falcons and their prey, houbara bustards and pigeons, possibly along the food chain. Western blotting detected specific anti-FalCoV UAE-HKU27 antibodies in 33 (75%) of 44 falcon serum samples, supporting genuine infection in falcons after virus acquisition. QuaCoV UAE-HKU30 belongs to the same CoV species as porcine coronavirus HKU15 (PorCoV HKU15) and sparrow coronavirus HKU17 (SpCoV HKU17), discovered previously from swine and tree sparrows, respectively, supporting avian-to-swine transmission. Recombination involving the spike protein is common among deltacoronaviruses, which may facilitate cross-species transmission. FalCoV UAE-HKU27, HouCoV UAE-HKU28, and PiCoV UAE-HKU29 originated from recombination between white-eye coronavirus HKU16 (WECoV HKU16) and magpie robin coronavirus HKU18 (MRCoV HKU18), QuaCoV UAE-HKU30 from recombination between PorCoV HKU15/SpCoV HKU17 and munia coronavirus HKU13 (MunCoV HKU13), and PorCoV HKU15 from recombination between SpCoV HKU17 and bulbul coronavirus HKU11 (BuCoV HKU11). Birds in the Middle East are hosts for diverse deltacoronaviruses with potential for interspecies transmission.IMPORTANCE During an attempt to explore the diversity of deltacoronaviruses among mammals and birds in Dubai, four novel deltacoronaviruses were detected in fecal samples from eight birds of four different species: FalCoV UAE-HKU27 from a falcon, HouCoV UAE-HKU28 from a houbara bustard, PiCoV UAE-HKU29 from a pigeon, and QuaCoV UAE-HKU30 from five quails. Genome analysis revealed evidence of recent interspecies transmission between falcons and their prey, houbara bustards and pigeons, possibly along the food chain, as well as avian-to-swine transmission. Recombination, which is known to occur frequently in some coronaviruses, was also common among these deltacoronaviruses and occurred predominantly at the spike region. Such recombination, involving the receptor binding protein, may contribute to the emergence of new viruses capable of infecting new hosts. Birds in the Middle East are hosts for diverse deltacoronaviruses with potential for interspecies transmission.
Keywords: Middle East; coronavirus; deltacoronavirus; falcon; houbara bustard; interspecies jumping; pigeon; quail.
Copyright © 2018 American Society for Microbiology.
Figures



Similar articles
-
Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus.J Virol. 2012 Apr;86(7):3995-4008. doi: 10.1128/JVI.06540-11. Epub 2012 Jan 25. J Virol. 2012. PMID: 22278237 Free PMC article.
-
Characterization of Deltacoronavirus in Black-Headed Gulls (Chroicocephalus ridibundus) in South China Indicating Frequent Interspecies Transmission of the Virus in Birds.Front Microbiol. 2022 May 12;13:895741. doi: 10.3389/fmicb.2022.895741. eCollection 2022. Front Microbiol. 2022. PMID: 35633699 Free PMC article.
-
Chicken or Porcine Aminopeptidase N Mediates Cellular Entry of Pseudoviruses Carrying Spike Glycoprotein from the Avian Deltacoronaviruses HKU11, HKU13, and HKU17.J Virol. 2023 Feb 28;97(2):e0194722. doi: 10.1128/jvi.01947-22. Epub 2023 Jan 19. J Virol. 2023. PMID: 36656013 Free PMC article.
-
Coronavirus diversity, phylogeny and interspecies jumping.Exp Biol Med (Maywood). 2009 Oct;234(10):1117-27. doi: 10.3181/0903-MR-94. Epub 2009 Jun 22. Exp Biol Med (Maywood). 2009. PMID: 19546349 Review.
-
Birds as reservoirs: unraveling the global spread of Gamma- and Deltacoronaviruses.mBio. 2024 Oct 16;15(10):e0232424. doi: 10.1128/mbio.02324-24. Epub 2024 Sep 4. mBio. 2024. PMID: 39230281 Free PMC article. Review.
Cited by
-
Antiviral performance of graphene-based materials with emphasis on COVID-19: A review.Med Drug Discov. 2021 Sep;11:100099. doi: 10.1016/j.medidd.2021.100099. Epub 2021 May 25. Med Drug Discov. 2021. PMID: 34056572 Free PMC article. Review.
-
Deltacoronavirus Evolution and Transmission: Current Scenario and Evolutionary Perspectives.Front Vet Sci. 2021 Feb 10;7:626785. doi: 10.3389/fvets.2020.626785. eCollection 2020. Front Vet Sci. 2021. PMID: 33681316 Free PMC article. Review.
-
The Remarkable Evolutionary Plasticity of Coronaviruses by Mutation and Recombination: Insights for the COVID-19 Pandemic and the Future Evolutionary Paths of SARS-CoV-2.Viruses. 2022 Jan 2;14(1):78. doi: 10.3390/v14010078. Viruses. 2022. PMID: 35062282 Free PMC article. Review.
-
Novel Bat Alphacoronaviruses in Southern China Support Chinese Horseshoe Bats as an Important Reservoir for Potential Novel Coronaviruses.Viruses. 2019 May 7;11(5):423. doi: 10.3390/v11050423. Viruses. 2019. PMID: 31067830 Free PMC article.
-
Swine coronaviruses (SCoVs) and their emerging threats to swine population, inter-species transmission, exploring the susceptibility of pigs for SARS-CoV-2 and zoonotic concerns.Vet Q. 2022 Dec;42(1):125-147. doi: 10.1080/01652176.2022.2079756. Vet Q. 2022. PMID: 35584308 Free PMC article. Review.
References
-
- de Groot RJ, Baker SC, Baric R, Enjuanes L, Gorbalenya AE, Holmes KV, Perlman S, Poon L, Rottier PJM, Talbot PJ, Woo PCY, Ziebuhr J. 2012. Family Coronaviridae, p 806–828. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier, San Diego, CA.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

