Microscopic and Molecular Evidence of the First Elasmobranch Adomavirus, the Cause of Skin Disease in a Giant Guitarfish, Rhynchobatus djiddensis
- PMID: 29764943
- PMCID: PMC5954223
- DOI: 10.1128/mBio.00185-18
Microscopic and Molecular Evidence of the First Elasmobranch Adomavirus, the Cause of Skin Disease in a Giant Guitarfish, Rhynchobatus djiddensis
Abstract
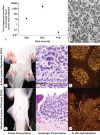
Only eight families of double-stranded DNA (dsDNA) viruses are known to infect vertebrate animals. During an investigation of papillomatous skin disease in an elasmobranch species, the giant guitarfish (Rhynchobatus djiddensis), a novel virus, distinct from all known viral families in regard to particle size, morphology, genome organization, and helicase phylogeny was discovered. Large inclusion bodies containing 75-nm icosahedral viral particles were present within epithelial cell nuclei in the proliferative skin lesions. Deep metagenomic sequencing revealed a 22-kb circular dsDNA viral genome, tentatively named guitarfish "adomavirus" (GAdoV), with only distant homology to two other fish viruses, Japanese eel endothelial cell-infecting virus (JEECV) and a recently reported marbled eel virus. Phylogenetic analysis of the helicase domain places the guitarfish virus in a novel clade that is equidistant between members of the Papillomaviridae and Polyomaviridae families. Specific PCR, quantitative PCR, and in situ hybridization were used to detect, quantify, and confirm that GAdoV DNA was localized to affected epithelial cell nuclei. Changes in the viral titer, as well as the presence of a hybridization signal, coincided with the progression and then final resolution of gross and microscopic lesions. The results indicate that GAdoV is the causative agent of the proliferative skin lesions.IMPORTANCE Cartilaginous fish, including the sharks and rays, evolved from ancestral fish species at least 400 million years ago. Even though they are the descendants of one of the most ancient vertebrate lineages, reports of viral diseases in these species are rare and poorly documented. Deep sequencing revealed a highly divergent virus, tentatively named guitarfish adomavirus, that is distantly related to known papillomaviruses and polyomaviruses. Out of the eight predicted viral genes, only the helicase could be identified as viral by sequence homology searches (BLAST), exemplifying the difficulties of discovering novel viruses within seas of unidentifiable "dark matter" associated with deep sequencing data. The novel adomavirus represents the first viral genome shown to cause clinical disease in a cartilaginous fish species, the giant guitarfish. Our findings demonstrate that emerging fish viruses are fertile ground to expand our understanding of viral evolution in vertebrates.
Keywords: adomavirus; cartilaginous fish; giant guitarfish; metagenomics; pathology; skin disease; virus discovery.
Copyright © 2018 Dill et al.
Figures
Similar articles
-
Virus-associated papillomatous skin lesions in a giant guitarfish Rhynchobatus djiddensis: a case report.Dis Aquat Organ. 2016 Jan 13;117(3):253-8. doi: 10.3354/dao02956. Dis Aquat Organ. 2016. PMID: 26758659
-
Novel adomavirus associated with proliferative skin lesions affecting the dermal denticles of a sand tiger shark (Carcharias taurus).Front Vet Sci. 2024 Oct 2;11:1470052. doi: 10.3389/fvets.2024.1470052. eCollection 2024. Front Vet Sci. 2024. PMID: 39415956 Free PMC article.
-
Complete Sequence of the Smallest Polyomavirus Genome, Giant Guitarfish (Rhynchobatus djiddensis) Polyomavirus 1.Genome Announc. 2016 May 19;4(3):e00391-16. doi: 10.1128/genomeA.00391-16. Genome Announc. 2016. PMID: 27198025 Free PMC article.
-
Evolutionary genomics of nucleo-cytoplasmic large DNA viruses.Virus Res. 2006 Apr;117(1):156-84. doi: 10.1016/j.virusres.2006.01.009. Epub 2006 Feb 21. Virus Res. 2006. PMID: 16494962 Review.
-
Evolution of the Large Nucleocytoplasmic DNA Viruses of Eukaryotes and Convergent Origins of Viral Gigantism.Adv Virus Res. 2019;103:167-202. doi: 10.1016/bs.aivir.2018.09.002. Epub 2018 Nov 10. Adv Virus Res. 2019. PMID: 30635076 Review.
Cited by
-
Draft Genome Sequence of an Adomavirus Associated with Raised Mucoid Skin Lesions on Smallmouth Bass (Micropterus dolomieu).Microbiol Resour Announc. 2020 Apr 2;9(14):e01479-19. doi: 10.1128/MRA.01479-19. Microbiol Resour Announc. 2020. PMID: 32241864 Free PMC article.
-
First Isolation of a Novel Aquatic Flavivirus from Chinook Salmon (Oncorhynchus tshawytscha) and Its In Vivo Replication in a Piscine Animal Model.J Virol. 2020 Jul 16;94(15):e00337-20. doi: 10.1128/JVI.00337-20. Print 2020 Jul 16. J Virol. 2020. PMID: 32434883 Free PMC article.
-
Global Organization and Proposed Megataxonomy of the Virus World.Microbiol Mol Biol Rev. 2020 Mar 4;84(2):e00061-19. doi: 10.1128/MMBR.00061-19. Print 2020 May 20. Microbiol Mol Biol Rev. 2020. PMID: 32132243 Free PMC article. Review.
-
Diversity, evolution, and emergence of fish viruses.J Virol. 2024 Jun 13;98(6):e0011824. doi: 10.1128/jvi.00118-24. Epub 2024 May 24. J Virol. 2024. PMID: 38785422 Free PMC article. Review.
-
Identification and characterization of a polyomavirus in the thornback skate (Raja clavata).Virol J. 2023 Aug 24;20(1):190. doi: 10.1186/s12985-023-02149-1. Virol J. 2023. PMID: 37620878 Free PMC article.
References
-
- Ng TF, Chen LF, Zhou Y, Shapiro B, Stiller M, Heintzman PD, Varsani A, Kondov NO, Wong W, Deng X, Andrews TD, Moorman BJ, Meulendyk T, MacKay G, Gilbertson RL, Delwart E. 2014. Preservation of viral genomes in 700-y-old caribou feces from a subarctic ice patch. Proc Natl Acad Sci U S A 111:16842–16847. doi:10.1073/pnas.1410429111. - DOI - PMC - PubMed
-
- Lauber C, Seitz S, Mattei S, Suh A, Beck J, Herstein J, Börold J, Salzburger W, Kaderali L, Briggs JAG, Bartenschlager R. 2017. Deciphering the origin and evolution of hepatitis B viruses by means of a family of non-enveloped fish viruses. Cell Host Microbe 22:387–399.e6. doi:10.1016/j.chom.2017.07.019. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
