Distinctive types of postzygotic single-nucleotide mosaicisms in healthy individuals revealed by genome-wide profiling of multiple organs
- PMID: 29763432
- PMCID: PMC5969758
- DOI: 10.1371/journal.pgen.1007395
Distinctive types of postzygotic single-nucleotide mosaicisms in healthy individuals revealed by genome-wide profiling of multiple organs
Abstract
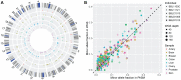
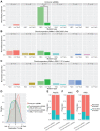
Postzygotic single-nucleotide mosaicisms (pSNMs) have been extensively studied in tumors and are known to play critical roles in tumorigenesis. However, the patterns and origin of pSNMs in normal organs of healthy humans remain largely unknown. Using whole-genome sequencing and ultra-deep amplicon re-sequencing, we identified and validated 164 pSNMs from 27 postmortem organ samples obtained from five healthy donors. The mutant allele fractions ranged from 1.0% to 29.7%. Inter- and intra-organ comparison revealed two distinctive types of pSNMs, with about half originating during early embryogenesis (embryonic pSNMs) and the remaining more likely to result from clonal expansion events that had occurred more recently (clonal expansion pSNMs). Compared to clonal expansion pSNMs, embryonic pSNMs had higher proportion of C>T mutations with elevated mutation rate at CpG sites. We observed differences in replication timing between these two types of pSNMs, with embryonic and clonal expansion pSNMs enriched in early- and late-replicating regions, respectively. An increased number of embryonic pSNMs were located in open chromatin states and topologically associating domains that transcribed embryonically. Our findings provide new insights into the origin and spatial distribution of postzygotic mosaicism during normal human development.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Postzygotic single-nucleotide mosaicisms in whole-genome sequences of clinically unremarkable individuals.Cell Res. 2014 Nov;24(11):1311-27. doi: 10.1038/cr.2014.131. Epub 2014 Oct 14. Cell Res. 2014. PMID: 25312340 Free PMC article.
-
Postzygotic single-nucleotide mosaicisms contribute to the etiology of autism spectrum disorder and autistic traits and the origin of mutations.Hum Mutat. 2017 Aug;38(8):1002-1013. doi: 10.1002/humu.23255. Epub 2017 May 30. Hum Mutat. 2017. PMID: 28503910 Free PMC article.
-
A model for postzygotic mosaicisms quantifies the allele fraction drift, mutation rate, and contribution to de novo mutations.Genome Res. 2018 Jul;28(7):943-951. doi: 10.1101/gr.230003.117. Epub 2018 Jun 6. Genome Res. 2018. PMID: 29875290 Free PMC article.
-
The ageing genome, clonal mosaicism and chronic disease.Curr Opin Genet Dev. 2017 Feb;42:8-13. doi: 10.1016/j.gde.2016.12.002. Epub 2017 Jan 6. Curr Opin Genet Dev. 2017. PMID: 28068559 Free PMC article. Review.
-
Somatic mosaicism: implications for disease and transmission genetics.Trends Genet. 2015 Jul;31(7):382-92. doi: 10.1016/j.tig.2015.03.013. Epub 2015 Apr 21. Trends Genet. 2015. PMID: 25910407 Free PMC article. Review.
Cited by
-
A rare case of a male child with post-zygotic de novo mosaic variant c.538C > T in MECP2 gene: a case report of Rett syndrome.BMC Neurol. 2021 Dec 2;21(1):469. doi: 10.1186/s12883-021-02500-5. BMC Neurol. 2021. PMID: 34856927 Free PMC article.
-
Genetic mosaicism in the human brain: from lineage tracing to neuropsychiatric disorders.Nat Rev Neurosci. 2022 May;23(5):275-286. doi: 10.1038/s41583-022-00572-x. Epub 2022 Mar 23. Nat Rev Neurosci. 2022. PMID: 35322263 Review.
-
Genomic mosaicism in the pathogenesis and inheritance of a Rett syndrome cohort.Genet Med. 2019 Jun;21(6):1330-1338. doi: 10.1038/s41436-018-0348-2. Epub 2018 Nov 8. Genet Med. 2019. PMID: 30405208 Free PMC article.
-
Early divergence of mutational processes in human fetal tissues.Sci Adv. 2019 May 29;5(5):eaaw1271. doi: 10.1126/sciadv.aaw1271. eCollection 2019 May. Sci Adv. 2019. PMID: 31149636 Free PMC article.
-
Parallel RNA and DNA analysis after deep sequencing (PRDD-seq) reveals cell type-specific lineage patterns in human brain.Proc Natl Acad Sci U S A. 2020 Jun 23;117(25):13886-13895. doi: 10.1073/pnas.2006163117. Epub 2020 Jun 10. Proc Natl Acad Sci U S A. 2020. PMID: 32522880 Free PMC article.
References
-
- Lupski JR. Genetics. Genome mosaicism—one human, multiple genomes. Science. 2013;341(6144):358–9. . - PubMed
-
- Poduri A, Evrony GD, Cai X, Walsh CA. Somatic mutation, genomic variation, and neurological disease. Science. 2013;341(6141):1237758 doi: 10.1126/science.1237758 . - DOI - PMC - PubMed
-
- Frank SA. Somatic mosaicism and disease. Curr Biol. 2014;24(12):R577–81. doi: 10.1016/j.cub.2014.05.021 . - DOI - PubMed
-
- Youssoufian H, Pyeritz RE. Mechanisms and consequences of somatic mosaicism in humans. Nat Rev Genet. 2002;3(10):748–58. doi: 10.1038/nrg906 . - DOI - PubMed
-
- Xu X, Yang X, Wu Q, Liu A, Yang X, Ye AY, et al. Amplicon Resequencing Identified Parental Mosaicism for Approximately 10% of "de novo" SCN1A Mutations in Children with Dravet Syndrome. Hum Mutat. 2015;36(9):861–72. Epub 2015/06/23. doi: 10.1002/humu.22819 . - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

