LlamaTags: A Versatile Tool to Image Transcription Factor Dynamics in Live Embryos
- PMID: 29754814
- PMCID: PMC6003873
- DOI: 10.1016/j.cell.2018.03.069
LlamaTags: A Versatile Tool to Image Transcription Factor Dynamics in Live Embryos
Abstract
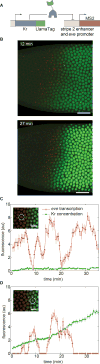
Embryonic cell fates are defined by transcription factors that are rapidly deployed, yet attempts to visualize these factors in vivo often fail because of slow fluorescent protein maturation. Here, we pioneer a protein tag, LlamaTag, which circumvents this maturation limit by binding mature fluorescent proteins, making it possible to visualize transcription factor concentration dynamics in live embryos. Implementing this approach in the fruit fly Drosophila melanogaster, we discovered stochastic bursts in the concentration of transcription factors that are correlated with bursts in transcription. We further used LlamaTags to show that the concentration of protein in a given nucleus heavily depends on transcription of that gene in neighboring nuclei; we speculate that this inter-nuclear signaling is an important mechanism for coordinating gene expression to delineate straight and sharp boundaries of gene expression. Thus, LlamaTags now make it possible to visualize the flow of information along the central dogma in live embryos.
Keywords: Drosophila melanogaster; Fushi-Tarazu; cell-to-cell communication; even-skipped; fluorescent protein maturation; live imaging; snail; transcription; transcription factors; translation.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos.Proc Natl Acad Sci U S A. 2014 Jul 22;111(29):10598-603. doi: 10.1073/pnas.1410022111. Epub 2014 Jul 3. Proc Natl Acad Sci U S A. 2014. PMID: 24994903 Free PMC article.
-
Using RNA Tags for Multicolor Live Imaging of Chromatin Loci and Transcription in Drosophila Embryos.Methods Mol Biol. 2020;2166:373-384. doi: 10.1007/978-1-0716-0712-1_22. Methods Mol Biol. 2020. PMID: 32710421 Free PMC article.
-
Mid-embryo patterning and precision in Drosophila segmentation: Krüppel dual regulation of hunchback.PLoS One. 2015 Mar 20;10(3):e0118450. doi: 10.1371/journal.pone.0118450. eCollection 2015. PLoS One. 2015. PMID: 25793381 Free PMC article.
-
The embryo as a laboratory: quantifying transcription in Drosophila.Trends Genet. 2014 Aug;30(8):364-75. doi: 10.1016/j.tig.2014.06.002. Epub 2014 Jul 6. Trends Genet. 2014. PMID: 25005921 Free PMC article. Review.
-
Lighting up the central dogma for predictive developmental biology.Curr Top Dev Biol. 2020;137:1-35. doi: 10.1016/bs.ctdb.2019.10.010. Epub 2019 Nov 22. Curr Top Dev Biol. 2020. PMID: 32143740 Review.
Cited by
-
Quantifying Temperature Compensation of Bicoid Gradients with a Fast T-Tunable Microfluidic Device.Biophys J. 2020 Sep 15;119(6):1193-1203. doi: 10.1016/j.bpj.2020.08.003. Epub 2020 Aug 12. Biophys J. 2020. PMID: 32853562 Free PMC article.
-
Enhancer Priming Enables Fast and Sustained Transcriptional Responses to Notch Signaling.Dev Cell. 2019 Aug 19;50(4):411-425.e8. doi: 10.1016/j.devcel.2019.07.002. Epub 2019 Aug 1. Dev Cell. 2019. PMID: 31378591 Free PMC article.
-
Notch-dependent and -independent transcription are modulated by tissue movements at gastrulation.Elife. 2022 May 18;11:e73656. doi: 10.7554/eLife.73656. Elife. 2022. PMID: 35583918 Free PMC article.
-
Chromatin expansion microscopy reveals nanoscale organization of transcription and chromatin.Science. 2023 Jul 7;381(6653):92-100. doi: 10.1126/science.ade5308. Epub 2023 Jul 6. Science. 2023. PMID: 37410825 Free PMC article.
-
Target gene responses differ when transcription factor levels are acutely decreased by nuclear export versus degradation.bioRxiv [Preprint]. 2024 May 20:2024.05.20.595009. doi: 10.1101/2024.05.20.595009. bioRxiv. 2024. Update in: Development. 2024 Nov 1;151(21):dev202775. doi: 10.1242/dev.202775. PMID: 38826476 Free PMC article. Updated. Preprint.
References
-
- Ay A, Knierer S, Sperlea A, Holland J, Ozbudak EM. Short-lived Her proteins drive robust synchronized oscillations in the zebrafish segmentation clock. Development. 2013;140:3244–3253. - PubMed
-
- Bieli D, Alborelli I, Harmansa S, Matsuda S, Caussinus E, Affolter M. Development and Application of Functionalized Protein Binders in Multicellular Organisms. Int Rev Cell Mol Biol. 2016;325:181–213. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

