An Iterative, Synthetic Approach To Engineer a High-Performance PhoB-Specific Reporter
- PMID: 29752265
- PMCID: PMC6029104
- DOI: 10.1128/AEM.00603-18
An Iterative, Synthetic Approach To Engineer a High-Performance PhoB-Specific Reporter
Abstract
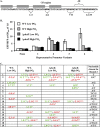
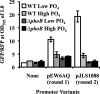
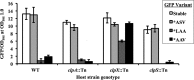
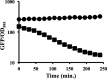
Transcriptional reporters are common tools for analyzing either the transcription of a gene of interest or the activity of a specific transcriptional regulator. Unfortunately, the latter application has the shortcoming that native promoters did not evolve as optimal readouts for the activity of a particular regulator. We sought to synthesize an optimized transcriptional reporter for assessing PhoB activity, aiming for maximal "on" expression when PhoB is active, minimal background in the "off" state, and no control elements for other regulators. We designed specific sequences for promoter elements with appropriately spaced PhoB-binding sites, and at 19 additional intervening nucleotide positions for which we did not predict sequence-specific effects, the bases were randomized. Eighty-three such constructs were screened in Vibrio fischeri, enabling us to identify bases at particular randomized positions that significantly correlated with high-level "on" or low-level "off" expression. A second round of promoter design rationally constrained 13 additional positions, leading to a reporter with high-level PhoB-dependent expression, essentially no background, and no other known regulatory elements. As expressed reporters, we used both stable and destabilized variants of green fluorescent protein (GFP), the latter of which has a half-life of 81 min in V. fischeri In culture, PhoB induced the reporter when phosphate was depleted to a concentration below 10 μM. During symbiotic colonization of its host squid, Euprymna scolopes, the reporter indicated heterogeneous phosphate availability in different light-organ microenvironments. Finally, testing this construct in other members of the Proteobacteria demonstrated its broader utility. The results illustrate how a limited ability to predict synthetic promoter-reporter performance can be overcome through iterative screening and reengineering.IMPORTANCE Transcriptional reporters can be powerful tools for assessing when a particular regulator is active; however, native promoters may not be ideal for this purpose. Optimal reporters should be specific to the regulator being examined and should maximize the difference between the "on" and "off" states; however, these properties are distinct from the selective pressures driving the evolution of natural promoters. Synthetic promoters offer a promising alternative, but our understanding often does not enable fully predictive promoter design, and the large number of alternative sequence possibilities can be intractable. In a synthetic promoter region with over 34 billion sequence variants, we identified bases correlated with favorable performance by screening only 83 candidates, allowing us to rationally constrain our design. We thereby generated an optimized reporter that is induced by PhoB and used it to explore the low-phosphate response of V. fischeri This promoter design strategy will facilitate the engineering of other regulator-specific reporters.
Keywords: Aliivibrio; photobacterium; synthetic biology.
Copyright © 2018 American Society for Microbiology.
Figures



Similar articles
-
An Expanded Transposon Mutant Library Reveals that Vibrio fischeri δ-Aminolevulinate Auxotrophs Can Colonize Euprymna scolopes.Appl Environ Microbiol. 2017 Feb 15;83(5):e02470-16. doi: 10.1128/AEM.02470-16. Print 2017 Mar 1. Appl Environ Microbiol. 2017. PMID: 28003196 Free PMC article.
-
The haem-uptake gene cluster in Vibrio fischeri is regulated by Fur and contributes to symbiotic colonization.Environ Microbiol. 2011 Nov;13(11):2855-64. doi: 10.1111/j.1462-2920.2011.02558.x. Epub 2011 Aug 30. Environ Microbiol. 2011. PMID: 21883801 Free PMC article.
-
New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ.Appl Environ Microbiol. 2006 Jan;72(1):802-10. doi: 10.1128/AEM.72.1.802-810.2006. Appl Environ Microbiol. 2006. PMID: 16391121 Free PMC article.
-
Control of biofilm formation and colonization in Vibrio fischeri: a role for partner switching?Environ Microbiol. 2010 Aug;12(8):2051-9. doi: 10.1111/j.1462-2920.2010.02269.x. Epub 2010 Jun 9. Environ Microbiol. 2010. PMID: 21966901 Free PMC article. Review.
-
Vibrio fischeri metabolism: symbiosis and beyond.Adv Microb Physiol. 2012;61:37-68. doi: 10.1016/B978-0-12-394423-8.00002-0. Adv Microb Physiol. 2012. PMID: 23046951 Review.
Cited by
-
Vibrio fischeri Possesses Xds and Dns Nucleases That Differentially Influence Phosphate Scavenging, Aggregation, Competence, and Symbiotic Colonization of Squid.Appl Environ Microbiol. 2022 Nov 22;88(22):e0163522. doi: 10.1128/aem.01635-22. Epub 2022 Nov 7. Appl Environ Microbiol. 2022. Corrected and republished in: Appl Environ Microbiol. 2024 Jun 18;90(6):e0032824. doi: 10.1128/aem.00328-24. PMID: 36342139 Free PMC article. Corrected and republished.
-
A lasting symbiosis: how Vibrio fischeri finds a squid partner and persists within its natural host.Nat Rev Microbiol. 2021 Oct;19(10):654-665. doi: 10.1038/s41579-021-00557-0. Epub 2021 Jun 4. Nat Rev Microbiol. 2021. PMID: 34089008 Free PMC article. Review.
-
Corrected and republished from: "Vibrio fischeri Possesses Xds and Dns Nucleases That Differentially Influence Phosphate Scavenging, Aggregation, Competence, and Symbiotic Colonization of Squid".Appl Environ Microbiol. 2024 Jun 18;90(6):e0032824. doi: 10.1128/aem.00328-24. Epub 2024 May 7. Appl Environ Microbiol. 2024. PMID: 38712952 Free PMC article.
-
Lighting the way: how the Vibrio fischeri model microbe reveals the complexity of Earth's "simplest" life forms.J Bacteriol. 2024 May 23;206(5):e0003524. doi: 10.1128/jb.00035-24. Epub 2024 May 2. J Bacteriol. 2024. PMID: 38695522 Free PMC article. Review.
-
Bacterial growth dynamics in a rhythmic symbiosis.Mol Biol Cell. 2024 Jun 1;35(6):ar79. doi: 10.1091/mbc.E24-01-0044. Epub 2024 Apr 10. Mol Biol Cell. 2024. PMID: 38598294 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

