Unraveling Haplotype Diversity of the Apical Membrane Antigen-1 Gene in Plasmodium falciparum Populations in Thailand
- PMID: 29742870
- PMCID: PMC5976018
- DOI: 10.3347/kjp.2018.56.2.153
Unraveling Haplotype Diversity of the Apical Membrane Antigen-1 Gene in Plasmodium falciparum Populations in Thailand
Abstract
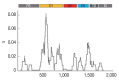
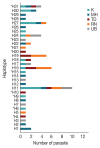
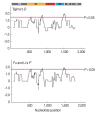
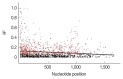
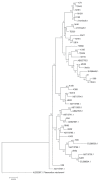
Development of an effective vaccine is critically needed for the prevention of malaria. One of the key antigens for malaria vaccines is the apical membrane antigen 1 (AMA-1) of the human malaria parasite Plasmodium falciparum, the surface protein for erythrocyte invasion of the parasite. The gene encoding AMA-1 has been sequenced from populations of P. falciparum worldwide, but the haplotype diversity of the gene in P. falciparum populations in the Greater Mekong Subregion (GMS), including Thailand, remains to be characterized. In the present study, the AMA-1 gene was PCR amplified and sequenced from the genomic DNA of 65 P. falciparum isolates from 5 endemic areas in Thailand. The nearly full-length 1,848 nucleotide sequence of AMA-1 was subjected to molecular analyses, including nucleotide sequence diversity, haplotype diversity and deduced amino acid sequence diversity and neutrality tests. Phylogenetic analysis and pairwise population differentiation (Fst indices) were performed to infer the population structure. The analyses identified 60 single nucleotide polymorphic loci, predominately located in domain I of AMA-1. A total of 31 unique AMA-1 haplotypes were identified, which included 11 novel ones. The phylogenetic tree of the AMA-1 haplotypes revealed multiple clades of AMA-1, each of which contained parasites of multiple geographical origins, consistent with the Fst indices indicating genetic homogeneity or gene flow among geographically distinct populations of P. falciparum in Thailand's borders with Myanmar, Laos and Cambodia. In summary, the study revealed novel haplotypes and population structure needed for the further advancement of AMA-1-based malaria vaccines in the GMS.
Keywords: Plasmodium falciparum; antigen; genetic polymorphism; human malaria; vaccine.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Genetic diversity of Plasmodium falciparum AMA-1 antigen from the Northeast Indian state of Tripura and comparison with global sequences: implications for vaccine development.Malar J. 2022 Feb 22;21(1):62. doi: 10.1186/s12936-022-04081-1. Malar J. 2022. PMID: 35193607 Free PMC article.
-
Genetic diversity and natural selection at the domain I of apical membrane antigen-1 (AMA-1) of Plasmodium falciparum in isolates from Iran.Exp Parasitol. 2012 Apr;130(4):456-62. doi: 10.1016/j.exppara.2012.01.006. Epub 2012 Jan 28. Exp Parasitol. 2012. PMID: 22306282
-
Natural selection and population genetic structure of domain-I of Plasmodium falciparum apical membrane antigen-1 in India.Infect Genet Evol. 2013 Aug;18:247-56. doi: 10.1016/j.meegid.2013.05.015. Epub 2013 Jun 6. Infect Genet Evol. 2013. PMID: 23747831
-
Genetic polymorphism in Plasmodium falciparum vaccine candidate antigens.Indian J Pathol Microbiol. 2005 Oct;48(4):429-38. Indian J Pathol Microbiol. 2005. PMID: 16366089 Review.
-
Natural selection on apical membrane antigen-1 of Plasmodium falciparum.Parassitologia. 1999 Sep;41(1-3):93-5. Parassitologia. 1999. PMID: 10697839 Review.
Cited by
-
Plasmodium falciparum Blood Stage Antimalarial Vaccines: An Analysis of Ongoing Clinical Trials and New Perspectives Related to Synthetic Vaccines.Front Microbiol. 2019 Dec 3;10:2712. doi: 10.3389/fmicb.2019.02712. eCollection 2019. Front Microbiol. 2019. PMID: 31849871 Free PMC article. Review.
-
Genotyping genetically heterogeneous Cyclospora cayetanensis infections to complement epidemiological case linkage.Parasitology. 2019 Sep;146(10):1275-1283. doi: 10.1017/S0031182019000581. Epub 2019 Jun 20. Parasitology. 2019. PMID: 31148531 Free PMC article.
-
Diversify and Conquer: The Vaccine Escapism of Plasmodium falciparum.Microorganisms. 2020 Nov 7;8(11):1748. doi: 10.3390/microorganisms8111748. Microorganisms. 2020. PMID: 33171746 Free PMC article. Review.
-
Genetic diversity of Plasmodium falciparum AMA-1 antigen from the Northeast Indian state of Tripura and comparison with global sequences: implications for vaccine development.Malar J. 2022 Feb 22;21(1):62. doi: 10.1186/s12936-022-04081-1. Malar J. 2022. PMID: 35193607 Free PMC article.
-
Reverse immunodynamics: a new method for identifying targets of protective immunity.Sci Rep. 2019 Feb 15;9(1):2164. doi: 10.1038/s41598-018-37288-x. Sci Rep. 2019. PMID: 30770839 Free PMC article.
References
-
- World Heath Organization . World Malaria Report, 2016. Geneva, Switzerland: World Heath Organization; 2016.
-
- Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, Sopha C, Chuor CM, Nguon C, Sovannaroth S, Pukrittayakamee S, Jittamala P, Chotivanich K, Chutasmit K, Suchatsoonthorn C, Runcharoen R, Hien TT, Thuy-Nhien NT, Thanh NV, Phu NH, Htut Y, Han KT, Aye KH, Mokuolu OA, Olaosebikan RR, Folaranmi OO, Mayxay M, Khanthavong M, Hongvanthong B, Newton PN, Onyamboko MA, Fanello CI, Tshefu AK, Mishra N, Valecha N, Phyo AP, Nosten F, Yi P, Tripura R, Borrmann S, Bashraheil M, Peshu J, Faiz MA, Ghose A, Hossain MA, Samad R, Rahman MR, Hasan MM, Islam A, Miotto O, Amato R, MacInnis B, Stalker J, Kwiatkowski DP, Bozdech Z, Jeeyapant A, Cheah PY, Sakulthaew T, Chalk J, Intharabut B, Silamut K, Lee SJ, Vihokhern B, Kunasol C, Imwong M, Tarning J, Taylor WJ, Yeung S, Woodrow CJ, Flegg JA, Das D, Smith J, Venkatesan M, Plowe CV, Stepniewska K, Guerin PJ, Dondorp AM, Day NP, White NJ. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371:411–423. - PMC - PubMed
-
- Manske M, Miotto O, Campino S, Auburn S, Almagro-Garcia J, Maslen G, O’Brien J, Djimde A, Doumbo O, Zongo I, Ouedraogo JB, Michon P, Mueller I, Siba P, Nzila A, Borrmann S, Kiara SM, Marsh K, Jiang H, Su XZ, Amaratunga C, Fairhurst R, Socheat D, Nosten F, Imwong M, White NJ, Sanders M, Anastasi E, Alcock D, Drury E, Oyola S, Quail MA, Turner DJ, Ruano-Rubio V, Jyothi D, Amenga-Etego L, Hubbart C, Jeffreys A, Rowlands K, Sutherland C, Roper C, Mangano V, Modiano D, Tan JC, Ferdig MT, Amambua-Ngwa A, Conway DJ, Takala-Harrison S, Plowe CV, Rayner JC, Rockett KA, Clark TG, Newbold CI, Berriman M, MacInnis B, Kwiatkowski DP. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature. 2012;487:375–379. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

