Mobilization of retrotransposons as a cause of chromosomal diversification and rapid speciation: the case for the Antarctic teleost genus Trematomus
- PMID: 29739320
- PMCID: PMC5941688
- DOI: 10.1186/s12864-018-4714-x
Mobilization of retrotransposons as a cause of chromosomal diversification and rapid speciation: the case for the Antarctic teleost genus Trematomus
Abstract
Background: The importance of transposable elements (TEs) in the genomic remodeling and chromosomal rearrangements that accompany lineage diversification in vertebrates remains the subject of debate. The major impediment to understanding the roles of TEs in genome evolution is the lack of comparative and integrative analyses on complete taxonomic groups. To help overcome this problem, we have focused on the Antarctic teleost genus Trematomus (Notothenioidei: Nototheniidae), as they experienced rapid speciation accompanied by dramatic chromosomal diversity. Here we apply a multi-strategy approach to determine the role of large-scale TE mobilization in chromosomal diversification within Trematomus species.
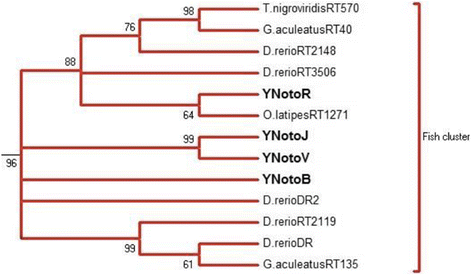
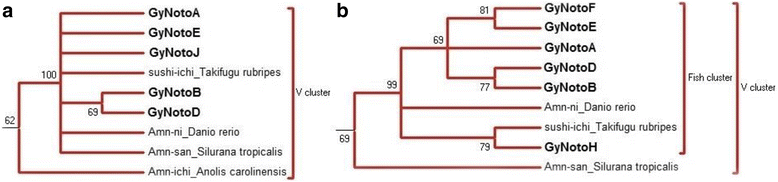
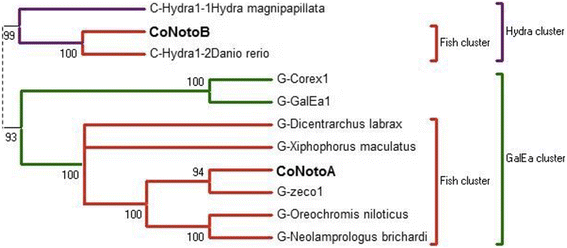
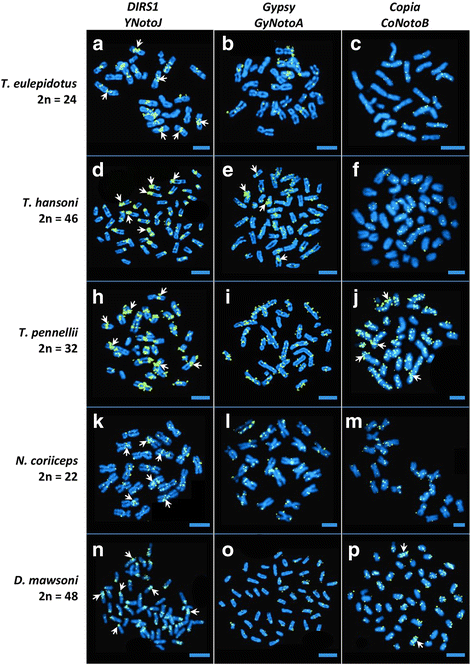
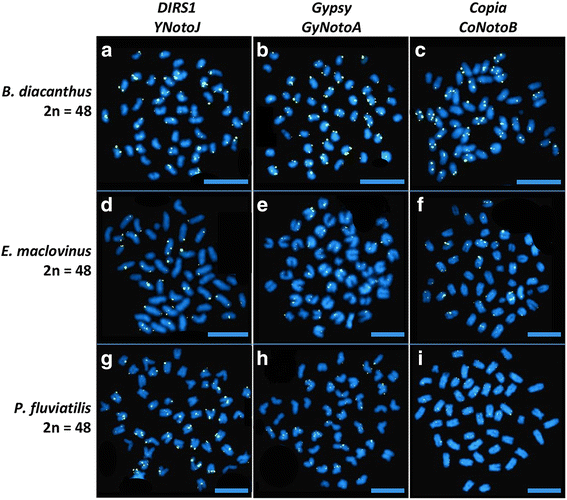
Results: Despite the extensive chromosomal rearrangements observed in Trematomus species, our measurements revealed strong interspecific genome size conservation. After identifying the DIRS1, Gypsy and Copia retrotransposon superfamilies in genomes of 13 nototheniid species, we evaluated their diversity, abundance (copy numbers) and chromosomal distribution. Four families of DIRS1, nine of Gypsy, and two of Copia were highly conserved in these genomes; DIRS1 being the most represented within Trematomus genomes. Fluorescence in situ hybridization mapping showed preferential accumulation of DIRS1 in centromeric and pericentromeric regions, both in Trematomus and other nototheniid species, but not in outgroups: species of the Sub-Antarctic notothenioid families Bovichtidae and Eleginopsidae, and the non-notothenioid family Percidae.
Conclusions: In contrast to the outgroups, High-Antarctic notothenioid species, including the genus Trematomus, were subjected to strong environmental stresses involving repeated bouts of warming above the freezing point of seawater and cooling to sub-zero temperatures on the Antarctic continental shelf during the past 40 millions of years (My). As a consequence of these repetitive environmental changes, including thermal shocks; a breakdown of epigenetic regulation that normally represses TE activity may have led to sequential waves of TE activation within their genomes. The predominance of DIRS1 in Trematomus species, their transposition mechanism, and their strategic location in "hot spots" of insertion on chromosomes are likely to have facilitated nonhomologous recombination, thereby increasing genomic rearrangements. The resulting centric and tandem fusions and fissions would favor the rapid lineage diversification, characteristic of the nototheniid adaptive radiation.
Keywords: Chomosomal rearrangements; DIRS1 insertion hot spots; FISH; Nototheniidae; Retrotransposons; Speciation; Trematomus.
Conflict of interest statement
Authors’ information
2: LP is in charge of the cytometers from the imaging platform of Sorbonne Université, 75,252 Paris, cedex 05
Ethics approval and consent to participate
Ethical approval for all procedures was granted by the ethics committee of the Ministère de l’Environnement and the French Polar Research Institute (Institut Paul Emile Victor – IPEV), which approved all our fieldwork. The experiments complied with the Code of Ethics of Animal Experimentation in the Antarctic sector.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Multiple independent chromosomal fusions accompanied the radiation of the Antarctic teleost genus Trematomus (Notothenioidei:Nototheniidae).BMC Evol Biol. 2020 Mar 20;20(1):39. doi: 10.1186/s12862-020-1600-3. BMC Evol Biol. 2020. PMID: 32192426 Free PMC article.
-
Insertion Hot Spots of DIRS1 Retrotransposon and Chromosomal Diversifications among the Antarctic Teleosts Nototheniidae.Int J Mol Sci. 2019 Feb 6;20(3):701. doi: 10.3390/ijms20030701. Int J Mol Sci. 2019. PMID: 30736325 Free PMC article.
-
Chromosomal distribution and evolution of abundant retrotransposons in plants: gypsy elements in diploid and polyploid Brachiaria forage grasses.Chromosome Res. 2015 Sep;23(3):571-82. doi: 10.1007/s10577-015-9492-6. Chromosome Res. 2015. PMID: 26386563
-
Eukaryote DIRS1-like retrotransposons: an overview.BMC Genomics. 2011 Dec 20;12:621. doi: 10.1186/1471-2164-12-621. BMC Genomics. 2011. PMID: 22185659 Free PMC article. Review.
-
Non-Antarctic notothenioids: Past phylogenetic history and contemporary phylogeographic implications in the face of environmental changes.Mar Genomics. 2016 Feb;25:1-9. doi: 10.1016/j.margen.2015.11.007. Epub 2015 Nov 21. Mar Genomics. 2016. PMID: 26610933 Review.
Cited by
-
Salmonidae Genome: Features, Evolutionary and Phylogenetic Characteristics.Genes (Basel). 2022 Nov 27;13(12):2221. doi: 10.3390/genes13122221. Genes (Basel). 2022. PMID: 36553488 Free PMC article. Review.
-
The complete mitochondrial genome of Trematomus hansoni Boulenger, 1902 (Perciformes, Nototheniidae).Mitochondrial DNA B Resour. 2024 Jun 3;9(6):701-706. doi: 10.1080/23802359.2024.2358959. eCollection 2024. Mitochondrial DNA B Resour. 2024. PMID: 38835641 Free PMC article.
-
Chromosome-Level Genome Assembly and Circadian Gene Repertoire of the Patagonia Blennie Eleginops maclovinus-The Closest Ancestral Proxy of Antarctic Cryonotothenioids.Genes (Basel). 2023 May 30;14(6):1196. doi: 10.3390/genes14061196. Genes (Basel). 2023. PMID: 37372376 Free PMC article.
-
Antarctic blackfin icefish genome reveals adaptations to extreme environments.Nat Ecol Evol. 2019 Mar;3(3):469-478. doi: 10.1038/s41559-019-0812-7. Epub 2019 Feb 25. Nat Ecol Evol. 2019. PMID: 30804520 Free PMC article.
-
Multiple independent chromosomal fusions accompanied the radiation of the Antarctic teleost genus Trematomus (Notothenioidei:Nototheniidae).BMC Evol Biol. 2020 Mar 20;20(1):39. doi: 10.1186/s12862-020-1600-3. BMC Evol Biol. 2020. PMID: 32192426 Free PMC article.
References
-
- Ozouf-Costaz C, Pisano E, Thaeron C, Hureau J-C. Antarctic fish chromosome banding: significance for evolutionary studies. Cybium. 1997;21:399–409.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

