Mtf2-PRC2 control of canonical Wnt signaling is required for definitive erythropoiesis
- PMID: 29736258
- PMCID: PMC5928144
- DOI: 10.1038/s41421-018-0022-5
Mtf2-PRC2 control of canonical Wnt signaling is required for definitive erythropoiesis
Abstract
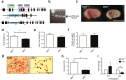
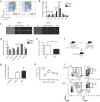
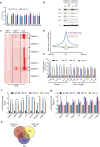
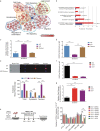
Polycomb repressive complex 2 (PRC2) accessory proteins play substoichiometric, tissue-specific roles to recruit PRC2 to specific genomic loci or increase enzymatic activity, while PRC2 core proteins are required for complex stability and global levels of trimethylation of histone 3 at lysine 27 (H3K27me3). Here, we demonstrate a role for the classical PRC2 accessory protein Mtf2/Pcl2 in the hematopoietic system that is more akin to that of a core PRC2 protein. Mtf2-/- erythroid progenitors demonstrate markedly decreased core PRC2 protein levels and a global loss of H3K27me3 at promoter-proximal regions. The resulting de-repression of transcriptional and signaling networks blocks definitive erythroid development, culminating in Mtf2-/- embryos dying by e15.5 due to severe anemia. Gene regulatory network (GRN) analysis demonstrated Mtf2 directly regulates Wnt signaling in erythroblasts, leading to activated canonical Wnt signaling in Mtf2-deficient erythroblasts, while chemical inhibition of canonical Wnt signaling rescued Mtf2-deficient erythroblast differentiation in vitro. Using a combination of in vitro, in vivo and systems analyses, we demonstrate that Mtf2 is a critical epigenetic regulator of Wnt signaling during erythropoiesis and recast the role of polycomb accessory proteins in a tissue-specific context.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
Tissue-Specific Tumour Suppressor and Oncogenic Activities of the Polycomb-like Protein MTF2.Genes (Basel). 2023 Sep 27;14(10):1879. doi: 10.3390/genes14101879. Genes (Basel). 2023. PMID: 37895228 Free PMC article. Review.
-
Mammalian polycomb-like Pcl2/Mtf2 is a novel regulatory component of PRC2 that can differentially modulate polycomb activity both at the Hox gene cluster and at Cdkn2a genes.Mol Cell Biol. 2011 Jan;31(2):351-64. doi: 10.1128/MCB.00259-10. Epub 2010 Nov 8. Mol Cell Biol. 2011. PMID: 21059868 Free PMC article.
-
MTF2 recruits Polycomb Repressive Complex 2 by helical-shape-selective DNA binding.Nat Genet. 2018 Jul;50(7):1002-1010. doi: 10.1038/s41588-018-0134-8. Epub 2018 May 28. Nat Genet. 2018. PMID: 29808031
-
A novel role of metal response element binding transcription factor 2 at the Hox gene cluster in the regulation of H3K27me3 by polycomb repressive complex 2.Oncotarget. 2018 May 29;9(41):26572-26585. doi: 10.18632/oncotarget.25505. eCollection 2018 May 29. Oncotarget. 2018. PMID: 29899877 Free PMC article.
-
The Role of Polycomb Proteins in Cell Lineage Commitment and Embryonic Development.Epigenomes. 2022 Aug 12;6(3):23. doi: 10.3390/epigenomes6030023. Epigenomes. 2022. PMID: 35997369 Free PMC article. Review.
Cited by
-
Polycomb function in early mouse development.Cell Death Differ. 2025 Jan;32(1):90-99. doi: 10.1038/s41418-024-01340-3. Epub 2024 Jul 12. Cell Death Differ. 2025. PMID: 38997437 Review.
-
DNA methylation drives hematopoietic stem cell aging phenotypes after proliferative stress.Geroscience. 2024 Oct 11. doi: 10.1007/s11357-024-01360-4. Online ahead of print. Geroscience. 2024. PMID: 39390312
-
Genetic Origins of the Two Canis lupus familiaris (Dog) Freight Dog Populations.J Hered. 2022 May 16;113(2):160-170. doi: 10.1093/jhered/esac002. J Hered. 2022. PMID: 35575082 Free PMC article.
-
Targeting the MTF2-MDM2 Axis Sensitizes Refractory Acute Myeloid Leukemia to Chemotherapy.Cancer Discov. 2018 Nov;8(11):1376-1389. doi: 10.1158/2159-8290.CD-17-0841. Epub 2018 Aug 16. Cancer Discov. 2018. PMID: 30115703 Free PMC article.
-
The roles of Polycomb repressive complexes in mammalian development and cancer.Nat Rev Mol Cell Biol. 2021 May;22(5):326-345. doi: 10.1038/s41580-021-00341-1. Epub 2021 Mar 15. Nat Rev Mol Cell Biol. 2021. PMID: 33723438 Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

